Introduction
For high-throughput profiling of miRNA expression
MicroRNAs (miRNAs) are small non-coding RNAs that regulate gene expression at the post-transcriptional level. Usually 21-23 nucleotides in length, miRNAs are important modulators in cellular pathways and are highly conserved in eukaryotic organisms. Irregularities in miRNA-regulated gene expression have been found to be associated with cancers, cardiovascular disorders and a variety of other diseases.
The miProfile miRNA PCR Arrays are designed for profiling the expression of pre-defined or customized sets of miRNAs in various tissues or cells. The resulting differential expression of profiled miRNAs help researchers to identify those miRNAs that are of biological significance and importance relevant to their research. Each 96-well plate contains up to 84 pairs of PCR primers (forward: miRNA-specific primer; reverse: universal primer) and 12 wells of different types of controls, which are used to monitor the efficiency of the entire experimental process – from reverse transcription to qPCR reaction.
Each miRNA-specific primer used in the qPCR arrays has been experimentally validated to yield a single dissociation curve peak and to generate a single amplification of the correct size for the targeted miRNA. A cDNA pool, containing reverse transcript products of 10 different tissue total RNAs, was used as the qPCR validation template.
Using a universal real-time PCR condition, one can easily profile and analyze the miRNA expression in a high-throughput fashion.The All-in-One™ miRNA First-Strand cDNA Synthesis Kits and qPCR Mix Kits are the recommended and supported RT-PCR reagents for use with the miProfile miRNA qPCR arrays. These reagents have been optimized to produce high sensitivity, efficiency, and specificity.
Key advantages
Validated miRNA primers
- Each miRNA primer is designed using a proprietary algorithm and experimentally validated
Robust performance
- Sensitive – Detect miRNA from as little as 10 pg of small RNA or 20 pg of total RNA
- Specific – Be able to distinguish miRNAs with single nucleotide mismatches. Each primer set has been experimental validated for specific amplification
- Broad linearity– Allow miRNA at varieties of expression level to be detected simultaneously
- Reproducible – High reproducibility (R2>0.99) for inter-array and intra-array replicates
Genome-wide coverage, pre-arranged groups, or customized groups
- Largest genome-wide miRNA coverage
- Cancer-related groups
- Customized miRNA arrays for focused study
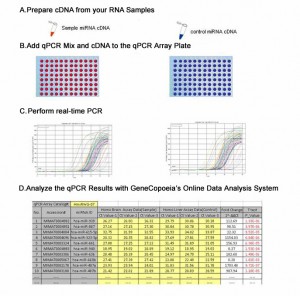 |
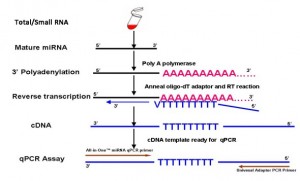 |
| Figure 1. miRNA PCR array experimental work flow (left) and miRNA RT-PCR mechanism (right). |
Performance data
|
Linear Range and Sensitivity (Total RNA)
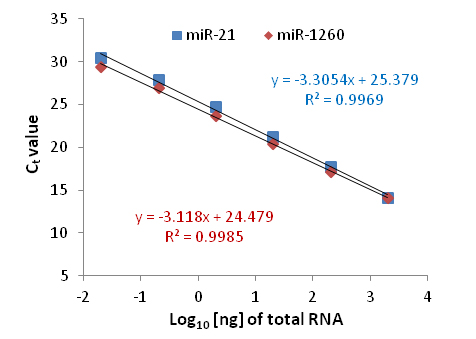
Figure 2. Broad linear range and high sensitivity
|
Inter-Array Reproducibility
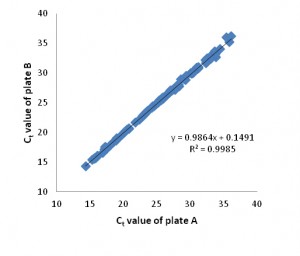
Figure 3. High inter-array reproducibility
|
|
Single Nucleotide Mismatches
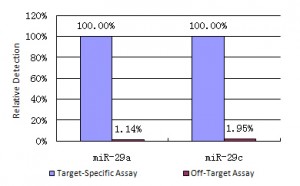
Figure 4. Specificity of miRNA detection
|
 |
Catalog Arrays
miProfile™ Catalog miRNA qPCR Arrays
miProfile™ Catalog miRNA qPCR Arrays profile the expression of pre-selected miRNAs, such as the whole genome, cancer-related or disease-related miRNAs. The cancer-related or dissease-related miRNAs are carefully chosen for their close correlation with a specific cancer or disease based on a thorough literature search of peer-reviewed publications.
GeneCopoeia offers the following catalog miRNA qPCR arrays:
Custom Arrays
miProfile™ Custom miRNA qPCR Arrays
For expression profiling of custom-specified miRNAs
miProfile™ Custom miRNA qPCR Arrays profile the expression of customer specified miRNAs. Each primer is designed using a proprietary algorithm and has been experimentally validated. Depending on the number of miRNAs and replicates to be analyzed, GeneCopoeia offers 14 layouts to choose from, including 6 types with 96-well plate and 8 types with 384-well plate. Different controls can also be included in the arrays to help monitor the sample quality, RT and PCR reaction efficiencies and replicates reproducibility:
- Spike-in reverse transcription control (RT): monitors the efficiency of the RT reaction.
- Positive PCR control (PCR): verifies the PCR efficiency.
- Housekeeping genes (HK): can be used as endogenous positive controls and for array normalization.
- Negative controls(NC): can be used as negative PCR control.
For the best performance, All-in-One™ miRNA First-Strand cDNA Synthesis Kit and All-in-One™ qPCR Mix are the recommended and supported reagents for use with these arrays.
Key advantages
|
 |
Validated primers
- Each primer is designed using a proprietary algorithm and has been experimentally validated
Flexible array design
- GeneCopoeia offers 14 layouts to choose from, including 6 types with 96-well plate and 8 types with 384-well plate.
Robust performance
- Sensitive – Detect miRNA from as little as 10 pg of small RNA or 20 pg of total RNA
- Specific – Be able to distinguish miRNAs with single nucleotide mismatches. Each primer has been experimental validated for specific amplification
- Broad linearity – Allow miRNA at varieties of expression level to be detected simultaneously
- Reproducible -High reproducibility (R2> 0.99) for inter-array and intra-array replicates
| Cat. No. |
Product name |
Species |
miRNAs |
Number of plates |
Document |
|
PAM-CS96 |
miProfile™ miRNA qPCR arrays custom services |
N/A |
Variable |
Variable |
Variable |












