GeneCopoeia’s Lentifect™ SARS-CoV-2 Spike-pseudotyped lentivirus products produce a specially-packaged version of standard lentivirus, whereby the vesicular stomatitis G (VSV-G) envelope glycoprotein is replaced by the Spike (S) protein from SARS-CoV-2, the virus responsible for the COVID-19 pandemic. These particles, also known as “pseudoviruses”, are typically used to infect cultured cells expressing ACE2, the receptor for SARS-CoV-2 on multiple tissue types (Figure 1).
Lentifect™ SARS-CoV-2 Spike-pseudotyped lentivirus and our ACE2- and TMPRSS2-expressing cell lines are great for several applications in a safe environment without the use of pathogenic SARS-CoV-2 viruses, such as:
- Assessment of the efficacy of vaccines against SARS-CoV-2 virus variants
- Studying the efficacy and mechanism of neutralizing antibodies against SARS-CoV-2 virus variants
- Development of antiviral therapeutic agents
- Studying the mechanism of virus-receptor interaction
Lentifect™ SARS-CoV-2 Spike-pseudotyped lentivirus products are part of GeneCopoeia’s COVID-19 Coronavirus Research Tools products. Note: Spike-pseudotyped lentiviruses are for research purposes only. They are not intended for diagnostic or treatment usage in humans.
Ordering information
Use the drop-down menus below to view products for lentivirus packaged with SARS-CoV-2 Spike protein.
For more information, please contact inquiry@genecopoeia.com.
GeneCopoeia’s Lentifect™ SARS-CoV-2 Spike protein-pseudotyped lentivirus is available as either premade, in vitro grade or custom-produced in vivo grade particles. Pseudoviral particles expressing Spike proteins from the original (“WT”) version as well as other variants, such as B.1.1.7 (first identified in the U.K.) and B.1.351 (first identified in South Africa) are available.
Advantages
- Accurate functional titers. Functional titers are measured by FACS sorting of ACE2-expressing HEK293T cells infected with GFP-expressing particles.
- Complete system. Premade, in vitro-grade purified particles can be used with GeneCopoeia’s ACE2-expressing HEK293T cells as well as other cells permissive to SARS-CoV-2 infection. Kits for do-it-yourself packaging of Spike-pseudotyped lentivirus also available.
- Quantitative measurements. Expression of GFP and luciferase enables quantitative determination of infection efficiency that is more accurate than traditional plaque reduction assays.
- Safe. Study SARS-CoV-2 Spike-mediated interactions with cells in the absence of pathogenic virus.
For more information, please contact inquiry@genecopoeia.com.
Lentifect™ lentiviral particles for SARS-CoV-2 coronavirus research (in vitro grade)
To view the Spike mutations in each pseudovirus variant, refer to the table “Spike variant mutational profiles” below.
1 The functional titers of SARS-CoV-2 Spike pseudotyped lentivirus are determined by FACS sorting of GFP positive ACE2-expressing ACE2/HEK293T cells (GeneCopoeia Cat.No. SL222) and by luciferase analysis (GeneCopoeia Cat.No. LF007).
2Titers of standard VSV-G pseudotyped lentivirus are determined by RT-qPCR to determine physical titer (copies/ml) and extrapolated to functional titer (TU/ml).
3The titer of the lentiviral particles without envelope protein is determined by RT-qPCR.
4SARS-CoV-2 strains carrying Spike amino acid mutant D614G have become, as of July, 2020, the prediminant global virus strains.
*Truncated SARS-CoV-2 Spike proteins carry a deletion of the 19 amino acid C-terminal endoplasmic reticulum retention signal.
Spike variant mutational profiles
At the time of product introduction, GeneCopoeia’s R&D team used the GISAID database to determine the most frequently-occurring mutations in the Spike proteins from each variant to incorporate into our pseudovirus particles. These mutational profiles are indicated below. Note that each variant can have different sub-lineages, which could contain a slightly different repertoire of mutations from the variants that we have made available. If you would like SARS-CoV-2 Spike-pseudotyped lentivirus expressing a different set of mutations than we have currently available. please contact inquiry@genecopoeia.com for a custom project.
| SARS-CoV-2 variant | Spike mutations |
|---|---|
| Wild type (catalog numbers SP001-100, SP101-100) | None |
| D614G (catalog numbers SP003-100, SP103-100) | D614G |
| N439K (catalog numbers SP004-100, SP104-100) | N439K |
| N439K & D614G (catalog numbers SP005-100, SP105-100) | N439K, D614G |
| B.1.1.7 (first identified in the U.K.; catalog numbers SP006-100, SP106-100) | Δ69-70, Δ144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H |
| B.1.351 (first identified in South Africa; catalog numbers SP007-100, SP107-100) | L18F, D80A, D215G,Δ242-244, K417N, E484K, N501Y, D614G, A701V |
| P.1 (first identified in Brazil; catalog numbers SP008-100, SP108-100) | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F |
| B.1.617.1 (first identified in India; catalog numbers SP009-100, SP109-100) | G142D, E154K, L452R, E484Q, D614G, P681R, Q1071H |
| B.1.427/B.1.429 (first identified in California, USA; catalog numbers SP010-100, SP110-100) | S13I, W152C, L452R, D614G |
| B.1.617.2 (first identified in India; catalog numbers SP011-100, SP111-100) | T19R, G142D, 156del, 157del, R158G, L452R, T478K, D614G, P681R, D950N |
| Lambda C.37 (first identified in Peru; catalog numbers SP012-100, SP112-100) | G75V, T76I, del246/253, L452Q, F490S, D614G, T859N |
| Mu B.1.621 (first identified in Columbia; catalog numbers SP013-100, SP113-100) | T95I, Y144S, Y145N, R346K, E484K, N501Y, D614G, P681H, D950N |
| Omicron B.1.1.529 (first identified in South Africa; catalog numbers SP015-100, SP115-100) | A67V, Del 69-70HV, T95I, G142D, Del 143-145VYY, N211I, del 212L, insert215-217 EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F |
| Omicron BA.2 (first identified in South Africa; catalog numbers SP016-100, SP116-100) | T19I, L24S, del25/27, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| Omicron BA.2.12.1 (first identified in South Africa; catalog numbers SP017-100, SP117-100) | T19I, L24S, del25/27, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452Q, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, S704L, N764K, D796Y, Q954H, N969K |
| Omicron BA.4/BA.5/BA.5.1 (first identified in South Africa; catalog numbers SP018-100, SP118-100) | T19I, L24S, del25/27, del69/70, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| Omicron BA.4.1 (first identified in South Africa; catalog numbers SP019-100, SP119-100) | V3G, T19I, L24S, del25/27, del69/70, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| Omicron BF.7 (first identified in South Africa; catalog numbers SP020-100, SP120-100) | T19I, L24S, del25/27, del69/70, G142D, V213G, G339D, R346T, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| Omicron BQ.1 (first identified in South Africa; catalog numbers SP021-100, SP121-100) | T19I, L24S, del25/27, del69/70, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, K444T, L452R, N460K, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| Omicron BQ.1.1 (first identified in South Africa; catalog numbers SP022-100, SP122-100) | T19I, L24S, del25/27, del69/70, G142D, V213G, G339D, R346T, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, K444T, L452R, N460K, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| Omicron XBB (first identified in South Africa; catalog numbers SP023-100, SP123-100) | T19I, L24S, del25/27, V83A, G142D, del144/144, H146Q, Q183E, V213E, G339H, R346T, L368I, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, V445P, G446S, N460K, S477N, T478K, E484A, F486S, F490S, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| Omicron XBB.1 (first identified in South Africa; catalog numbers SP024-100, SP124-100) | T19I, L24S, del25/27, V83A, G142D, del144/144, H146Q, Q183E, V213E, G252V, G339H, R346T, L368I, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, V445P, G446S, N460K, S477N, T478K, E484A, F486S, F490S, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| Omicron XBB.1.5 (first identified in South Africa; catalog numbers SP025-100, SP125-100) | T19I, L24S, del25/27, V83A, G142D, del144/144, H146Q, Q183E, V213E, G252V, G339H, R346T, L368I, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, V445P, G446S, N460K, S477N, T478K, E484A, F486P, F490S, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
COVID-19 Coronavirus Receptor Stable Cell Lines
For more information or to request a quote, please contact inquiry@genecopoeia.com.
Lenti-Pac™ lentivirus packaging kits for SARS-CoV-2 coronavirus research
GeneCopoeia manufactures kits that allow researchers to produce SARS-CoV-2 Spike-pseudotyped lentivirus, which you can order from the table below.
To view the Spike mutations in each pseudovirus variant, refer to the table “Spike variant mutational profiles” below.
Spike variant mutational profiles
At the time of product introduction, GeneCopoeia’s R&D team used the GISAID database to determine the most frequently-occurring mutations in the Spike proteins from each variant to incorporate into our pseudovirus packaging kits. These mutational profiles are indicated below. Note that each variant can have different sub-lineages, which could contain a slightly different repertoire of mutations from the variants that we have made available. If you would like SARS-CoV-2 Spike-pseudotyped lentivirus packaging kits expressing a different set of mutations than we have currently available. please contact inquiry@genecopoeia.com for a custom project.
| SARS-CoV-2 variant | Spike mutations |
| Wild type (catalog number LT011, LT021) | None |
| D614G (catalog number LT015, LT017) | D614G |
| B.1.1.7 (first identified in the U.K.; catalog number LT041) | Δ69-70, Δ144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H |
| B.1.351 (first identified in South Africa; catalog number LT044) | L18F, D80A, D215G,Δ242-244, K417N, E484K, N501Y, D614G, A701V |
| P.1 (first identified in Brazil; catalog number LT047) | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F |
For more information or to request a quote, please contact inquiry@genecopoeia.com.
GeneCopoeia has produced HEK293T cell lines expressing human ACE2 alone, or ACE2 in combination with TMPRSS2, a protease that promotes Spike-ACE2 receptor interaction. These cell lines express ACE2 and/or TMPRRSS2 mRNA at high levels (Figure 1). Due to display of ACE2 protein (Figure 2) and/or TMPRSS2 protein ((Figure 3) on the cell surface, these cells are amenable to infection by viruses expressing SARS-CoV-2 Spike protein in their envelopes.
You can order these cell lines from the table below. To obtain the datasheet for each cell line, please click on the designated catalog number.
In addition to the products below, custom stable cell lines are also available. For more information, please contact inquiry@genecopoeia.com.
For more information or to request a quote, please contact inquiry@genecopoeia.com.
Performance data
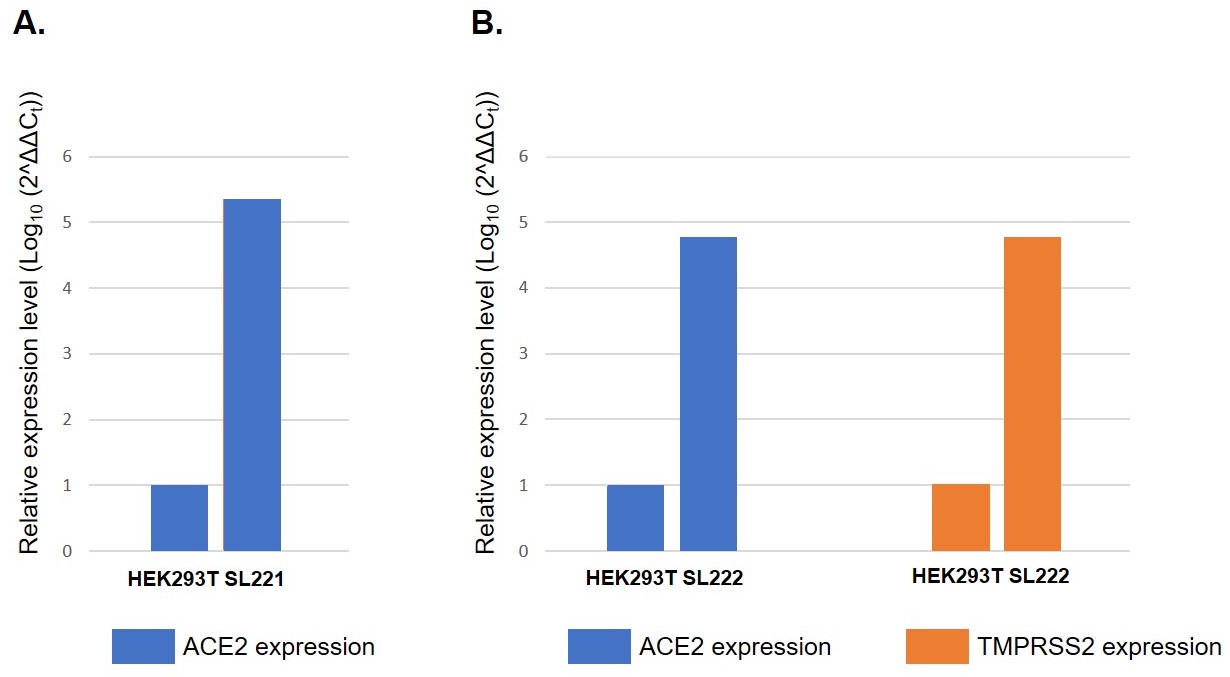
Figure 1. Relative mRNA expression levels of ACE2 and TMPRSS2 in GeneCopoeia stable cell lines. mRNA was amplified via RT-qPCR using primers specific for each gene. Ct values of ACE2 and TMPRSS2 were normalized to Ct values for housekeeping gene GAPDH. Relative expression levels were calculated from 2^ΔΔCt. A. ACE2 mRNA expression in parental cell line HEK293T and ACE2-overexpressing HEK293T cell line SL221. B. ACE2 and TMPRSS2 mRNA expression in parental cell line HEK293T and ACE2- and TMPRSS2-overexpressing cell line SL222.
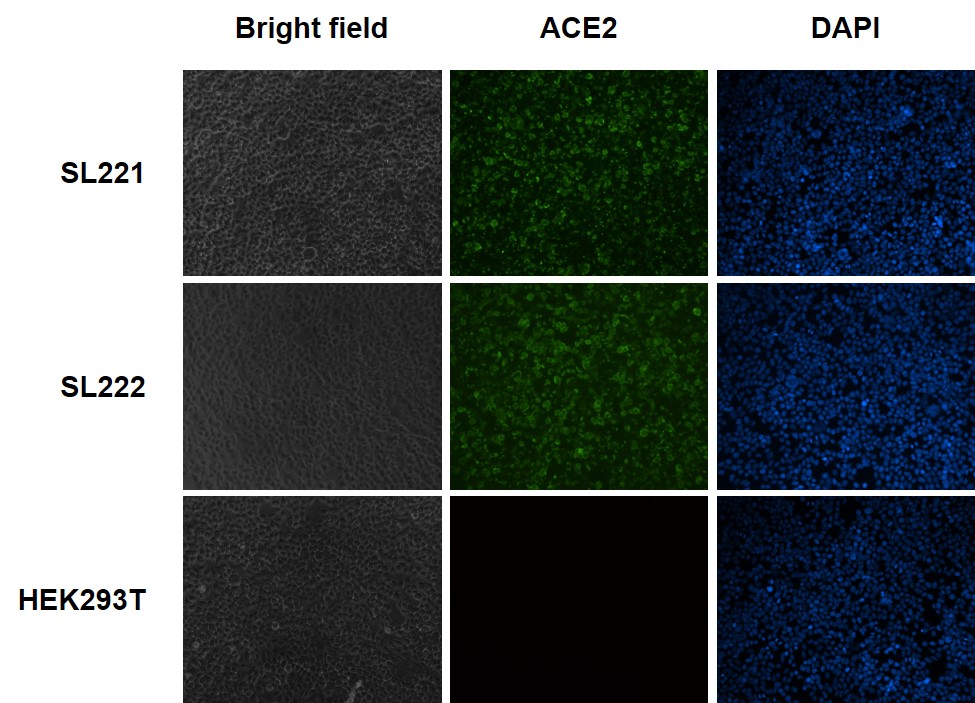
Figure 2. Immunofluorescence (IF) analysis of ACE2 protein expression on the cell surface of ACE2-expressing HEK293T cells (Cat.# SL221), ACE2/TMPRSS2 expressing HEK293T cells (Cat.# SL222), and HEK293T cells. Unpermeablized cells were fixed, blocked and incubated with the primary anti-ACE2 antibody, then washed and incubated with fluorescent-labeled secondary antibody, washed again, and stained with DAPI.
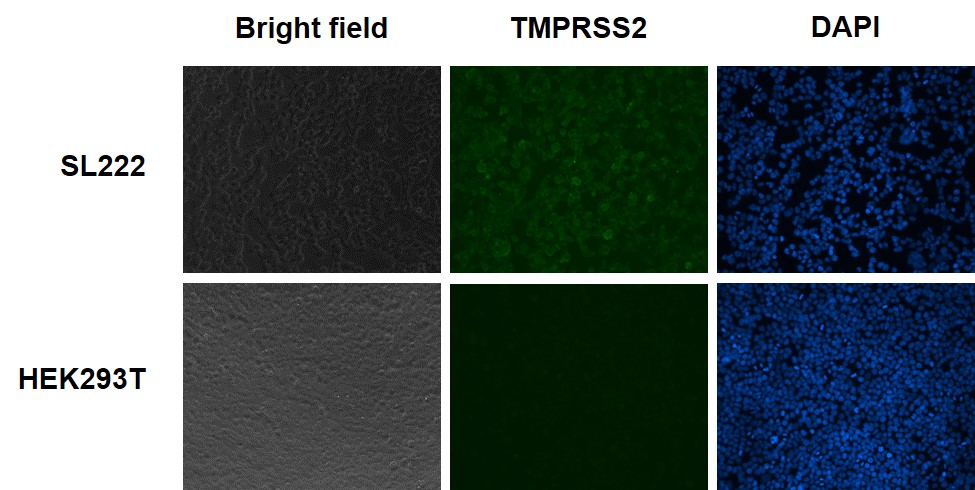
Figure 3. Immunofluorescence (IF) analysis of TMPRSS2 protein expression on the cell surface of TMPRSS2-expressing HEK293T cells (Cat.# SL222), and HEK293T cells. Unpermeablized cells were fixed, blocked and incubated with the primary anti-TMPRSS2 antibody, then washed and incubated with fluorescent-labeled secondary antibody, washed again, and stained with DAPI.
Performance data
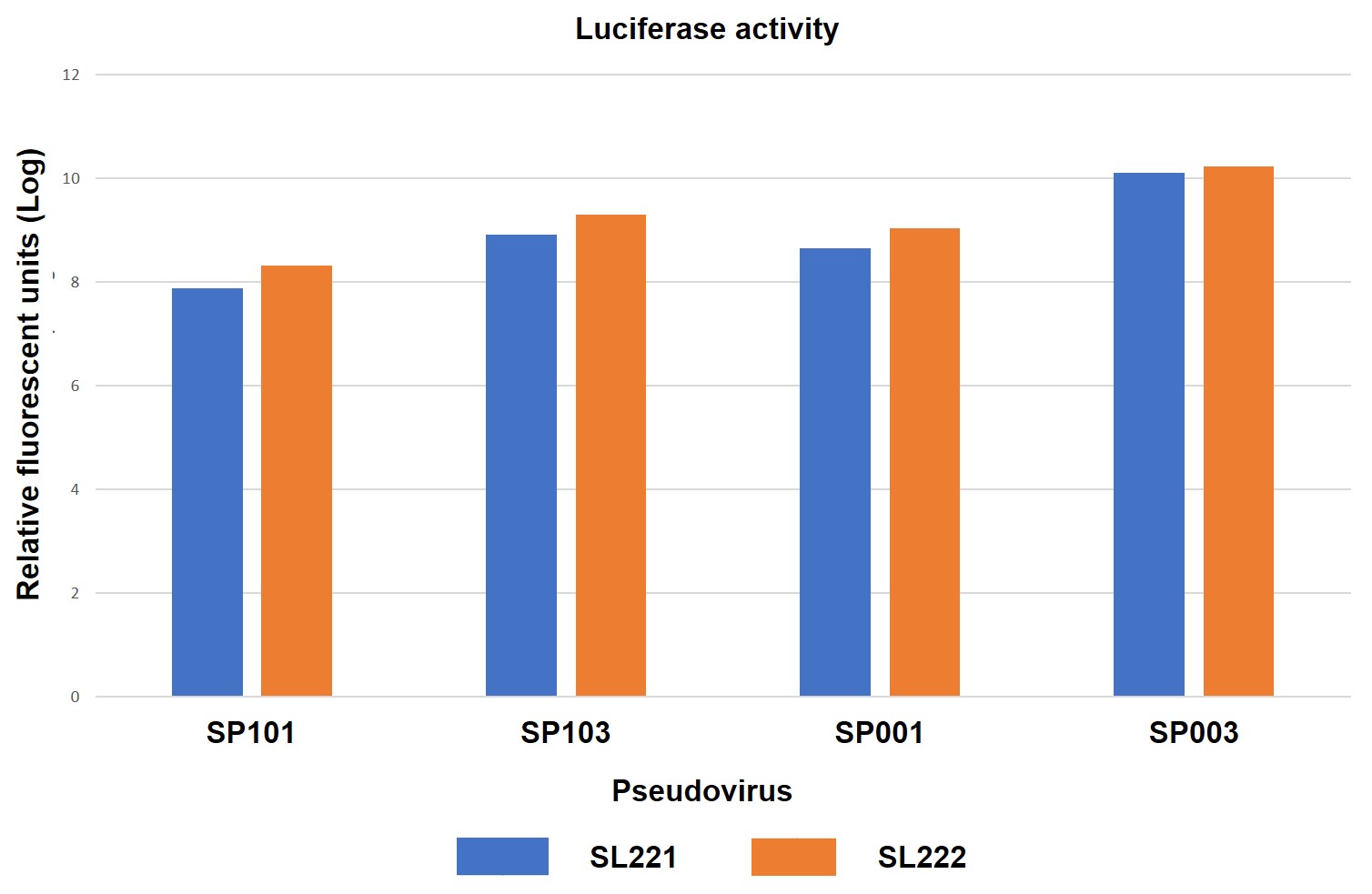
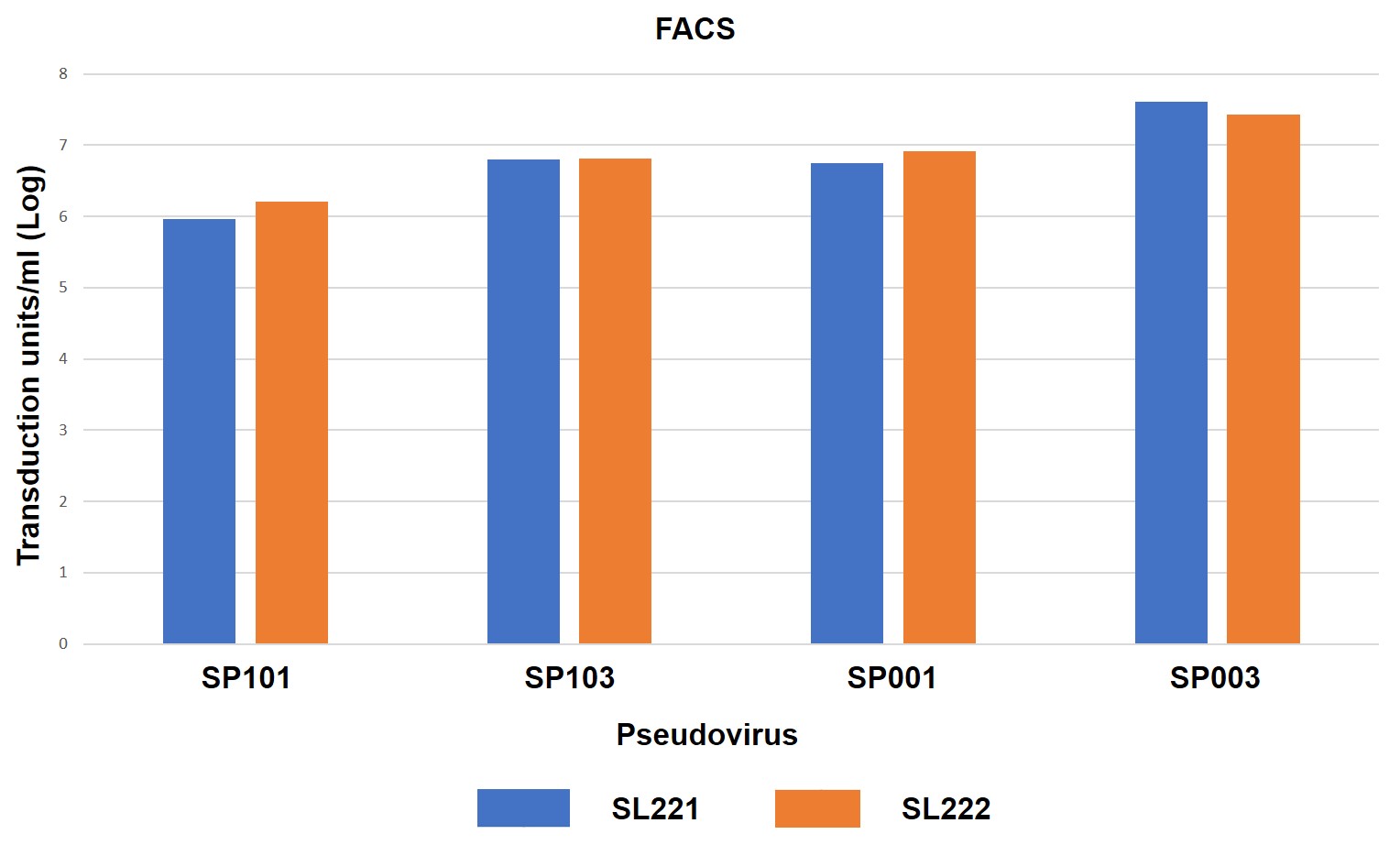
GeneCopoeia’s R&D team determined the ability of SARS-CoV-2 Spike-Pseudotyped Lentiviruses to infect ACE2-expressing cells, as shown in Figure 4.
A. |
B. |
C. |
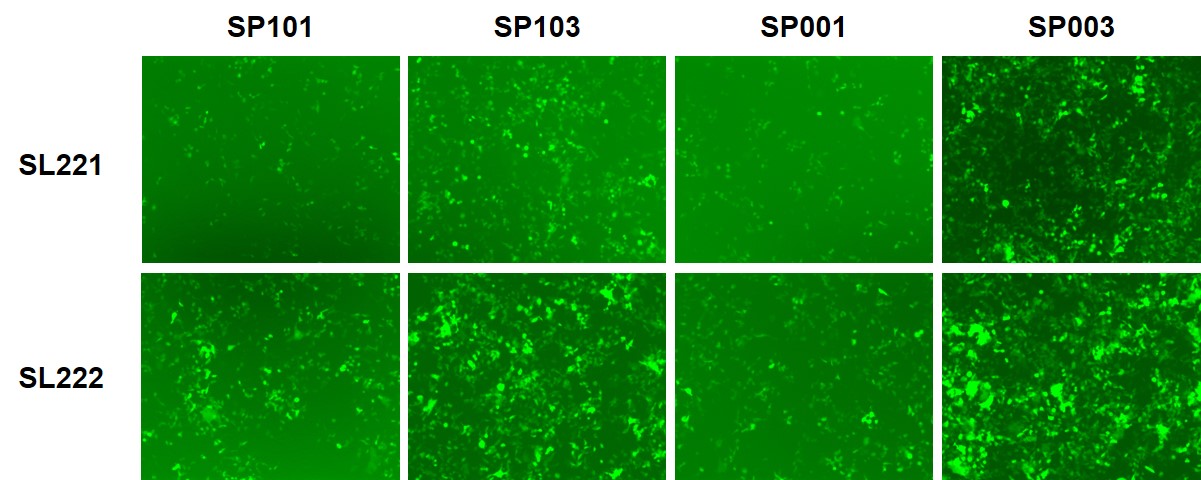
Figure 4. ACE2- or ACE2/TMPRSS2-expressing HEK293T cells transduced by eGFP/hLuc-expressing SARS-CoV-2 Spike pseudotyped lentiviruses (pseudoviruses). 10 μl of pseudovirus was used to transduce each cell line in a 96-well plate. 72 hours after transduction, cells were photographed under fluorescence microscopy, followed by measurement of luciferase activities or FACS sorting. SL221: HEK293T cells expressing ACE2. SL222: HEK293T cells expressing ACE2 and TMPRSS2. SP101: Full-length Spike protein, D614 variant. SP103: Full-length Spike protein, D614G variant. SP001: Truncated Spike protein, D614 variant. SP003: Truncated Spike protein, D614G variant. A. Fluorescence microscopy. B. Luciferase activity assay. 20 μl of cell lysate from pseudovirus transduced cell cultures were analyzed for firefly luciferase activity using GeneCopoeia’s Luc-Pair™ Firefly Luciferase HS Assay Kit (cat. no. LF007). C. FACS sorting for determination of titer in transduction units/ml.
Related products and services
Webinars
Title: Advanced Tools For COVID-19 Coronavirus Research From GeneCopoeia
Presented Wednesday, September 9, 2020
The COVID-19 global pandemic, caused by the SARS-CoV-2 coronavirus, highlights an urgent need for diagnostic reagents, antiviral therapeutics, and vaccine development. In this webinar, we will discuss GeneCopoeia’s advanced solutions including testing kits, SARS-CoV-2 pseudotyped virus, and other tools needed for research into combating this deadly disease.
Watch recorded webinar / Download slides
Publications
View selected publications citing GeneCopoeia’s SARS-CoV-2 Spike-pseudotyped lentivirus products from the recent literature
2022
- Dai, H., et al. (2022). Rapid differentiation of SARS-CoV-2 variants on a nanotechnology enhanced DNA-chip. Reseearch Square doi: 10.21203/rs.3.rs-1641024/v1 [SARS-CoV-2 Spike-pseudotyped lentiviral particles]
- Travis, B.J., et al. (2022). Significance of chlorine-dioxide-based oral rinses in preventing SARS-CoV-2 cell entry. Oral Diseases doi: 10.1111/odi.14319 [Lentifect™ SARS-CoV2 Spike-pseudotyped Lentiviral particles]
2021
- Tito, A., et al. (2021). Pomegranate Peel Extract as an Inhibitor of SARS-CoV-2 Spike Binding to Human ACE2 Receptor (in vitro): A Promising Source of Novel Antiviral Drugs. Frontiers in Chemistry doi: 10.3389/fchem.2021.638187 [SARS-CoV-2 Spike-pseudotyped lentiviral particles]
- Paidi, R.K., et al. (2021). Eugenol, a Component of Holy Basil (Tulsi) and Common Spice Clove, Inhibits the Interaction Between SARS-CoV-2 Spike S1 and ACE2 to Induce Therapeutic Responses. Journal of Neuroimmune Pharmacology doi: 10.1007/s11481-021-10028-1 [SARS-CoV-2 Spike-pseudotyped lentiviral particles]
- Basu, A., et al. (2021). A structurally conserved RNA element within SARS-CoV-2 ORF1a RNA and S mRNA regulates translation in response to viral S protein-induced signaling in human lung cells. Journal of Virology doi: 10.1128/JVI.01678-21 [SARS-CoV-2 Spike-pseudotyped lentiviral particles]
- Paidi, R.K., et al. (2021). Selective Inhibition of the Interaction between SARS-CoV-2 Spike S1 and ACE2 by SPIDAR Peptide Induces Anti-Inflammatory Therapeutic Responses. Journal of Immunology doi: 10.4049/jimmunol.2100144 [Lentifect™ SARS-CoV2 Spike-pseudotyped Lentiviral particles]
Frequently Asked Questions
Answer: TU/ml (transduction units per ml) is a common titration standard for lentivirus. It is a functional titer that represents the number of infectious virus particles, which is lower than the number of physical copies, in a sample. We used GFP expression from infection of our HEK293T cell line expressing ACE2 (catalog number SL221), the receptor for the SARS-CoV-2 virus. After infection, we used fluorescence activated cell sorting (FACS) to count the number of infected cells. Because this titer was determined by FACS, it is a digital determination of titer.
The RLU/ml titer is a functional titer based on firefly luciferase relative luminescence units/ml, similar to what was shown in a published paper (Ou, X., et al., 2020. Nature Communications doi: 10.1038/s41467-020-15562-9). This is dependent on the type of reagents used for the luciferase assay, however. We measured RLU/ml using our High Sensitivity Firefly Luciferase Kit, but one would expect the RLU titer to be lower using our standard luciferase kit with lower sensitivity. Therefore, it’s more meaningful to compare RLUs between the spike-pseudotyped virus vs. VSV-G virus using the same reagents at the same time.
Answer: The mix contains 3 plasmids. One of these plasmids expresses rev, and one of the plasmids expresses gag and pol. The 3rd plasmid expresses the envelope protein. This mix is identical to GeneCopoeia’s Lenti-Pac™ lentiviral packaging mix except 1) The VSV-G gene of the Lenti-Pac™ lentivirus packaging kit expressed on the envelope plasmid has been replaced with the Spike protein-expressng gene of SARS-CoV-2; and 2) the mix is formulated in a proprietary ratio.
Answer: The packaging mix is not intended to be used for amplification and extraction from bacteria. You could do it, but then we could not guarantee that you would get colonies expressing all 3 plasmids. In addition, you would likely not get the correct ratios needed among all 3 to get efficient packaging. We have determined this proprietary ratio for optimal virus packaging. Therefore, if you run out of packaging mix DNA, you would need to order more from GeneCopoeia. However, you can transform bacteria with the transfer plasmid, the plasmid that gets packaged by the mix, because this is a single plasmid.
Answer: No. The GFP control plasmid is included so that you know the kit is functioning properly. You can use the kit to package any 3rd-generation lentiviral expression plasmid, also referred to as the “transfer plasmid”. GeneCopoeia provides many clones that express ORFs, shRNAs, miRNAs and miRNA inhibitors, gene promoters, and CRISPR sgRNAs in more than 80 lentiviral vector backbones. For more information, visit our lentiviral clone page, or contact inquiry@genecopoeia.com.