Autoimmune diseases occur when the immune system, which normally does not attack a person’s own components, produces self-reacting T-cells or autoantibodies that recognize host self-components (proteins, nucleic acids, lipids, etc.), also known as autoantigens. Over 100 autoimmune diseases have been described, creating a critical need for tools that can detect large numbers of autoantibodies with high sensitivity.
GeneCopoeia’s OmicsArray™ Systemic Autoimmune-associated Antigen Array (Cat#: PA001) is a protein microarray enabling powerful detection of autoantibodies associated with many autoimmune diseases. The array carries 128 superior-quality purified antigens and internal controls spotted in duplicates onto nitrocellulose filters, which are adhered to glass slides. Antigens known to be associated with specific autoimmune diseases are chosen based on a thorough review of peer-reviewed publications. We have two versions of PA001 for customer to choose. PA001-V3 is a modified version based on PA001-V2 with 19 different antigens which are mostly associated with inflammatory myositis. Click here to download the antigen comparison of PA001-V2 and PA001-V3.
In addition to premade arrays, arrays containing customized sets of antigens are available, as well as array processing kits, array profiling services and data analysis. To order premade or custom arrays, please contact us.
GeneCopoeia’s OmicsArray™ Systemic Autoimmune-associated Antigen Array is part of the GeneCopoeia OmicsArray™ Antigen Microarray family.
Premade Antigen Arrays
The listed chip prices apply to academic customers. For custom arrays and services pricing, please contact us.
There are 19 different antigens between V2 and V3. The V3 contains more antigens associated with inflammatory myositis.| User Manual |
Prices are for customers from the U.S. and Canada only. For other customers, please contact your local distributor for details.
Antigen microarray processing kits and accessories
| User Manual | SDS |
Prices are for customers from the U.S. and Canada only. For other customers, please contact your local distributor for details.
Custom services
GeneCopoeia offers custom antigen microarray services in the following areas:- Custom array printing. GeneCopoeia will create custom antigen microarrays built to your specifications.
- Sample processing. Send us your blood, plasma, tissue, or other biological sample and we will prepare it for processing and incubation with any of our premade antigen microarrays or custom-built antigen microarrays for autoantibody profiling and other applications. For information on sample types to submit, consult the FAQ
- Data analysis. Once samples are processed and incubated with an antigen microarray, we will analyze the raw data. The standard analysis service includes: 1) An Excel file of the Net Signal Intensity (NSI) for each antigen on the array, normalized to internal controls; and 2) a heat map
⇒Return to the main OmicsArray™ Antigen Microarray page
- Largest collection of pre-made whole-protein antigen microarrays on the market.
- Largest number of whole-protein antigens specifically focused on autoimmune diseae research.
- Best combination of number of antigens per array (up to 120) with number of samples that can be processed per slide (up to 15).
Technology overview
GeneCopoeia’s OmicsArray™ antigen microarrays contain up to 120 purified proteins spotted onto nitrocellulose filters, which are adhered to glass slides. In addition, 8 spots are included for normalization. Each slide carries 16 identical arrays, and so can be used to process up to 15 samples simultaneously as well as a negative control. As little as 1 ul serum or 50 ul of other bio fluids are needed for each sample.
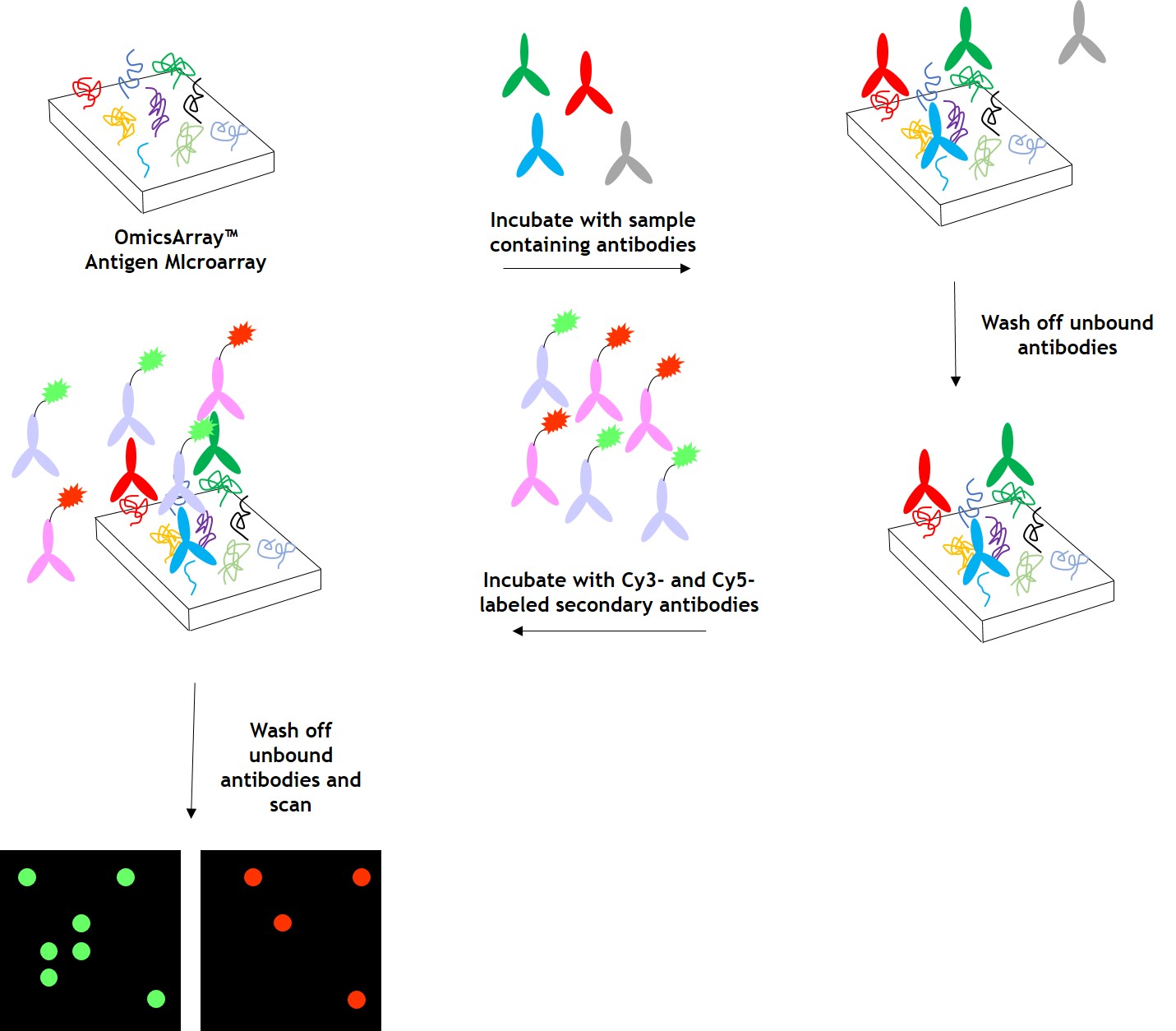
As shown in Figure 1, arrays are incubated with patient samples, and any antibodies in the samples bind to their cognate antigens on the array. The arrays are washed to remove unbound antibodies and other proteins, then co-incubated with Cy3- and Cy5-labeled secondary antibodies. The dual labeling strategy is intended to distinguish between immunoglobulin (Ig) subtypes present within samples. For example. a Cy3-labeled anti-IgG secondary antibody is used to detect IgG antibodies, and a Cy5-labeled anti-IgM secondary antibody is used to detect IgM antibodies. Fluorophore-labeled secondary antibodies are available for detecting IgA, IgD, IgE, IgG and IgM immunoglubulins, as well as IgG subclasses IgG1, IgG2, IgG3, and IgG4.
After washing to remove unbound secondary antibodies, signals are detected using a microarray scanner (e.g., GenePix® 4000B, InnoScan 710, or equivalents). The raw data is then be analyzed using GenePix® Pro 7.0, or Mapix software.
Figure 1. Workflow for detection of antibodies in samples using GeneCopoeia’s OmicsArray™ antigen microarrays.
Data analysis
GeneCopoeia provides data analysis as part of its custom antigen microarray services. After array scanning, raw data are collected and analyzed using GenePix® 7.0 software. The standard service includes data normalization to include fold changes of, for example, disease vs. healthy control, and a heat map showing each antigen ranked by level of fold change. Our service undergoes the following steps:
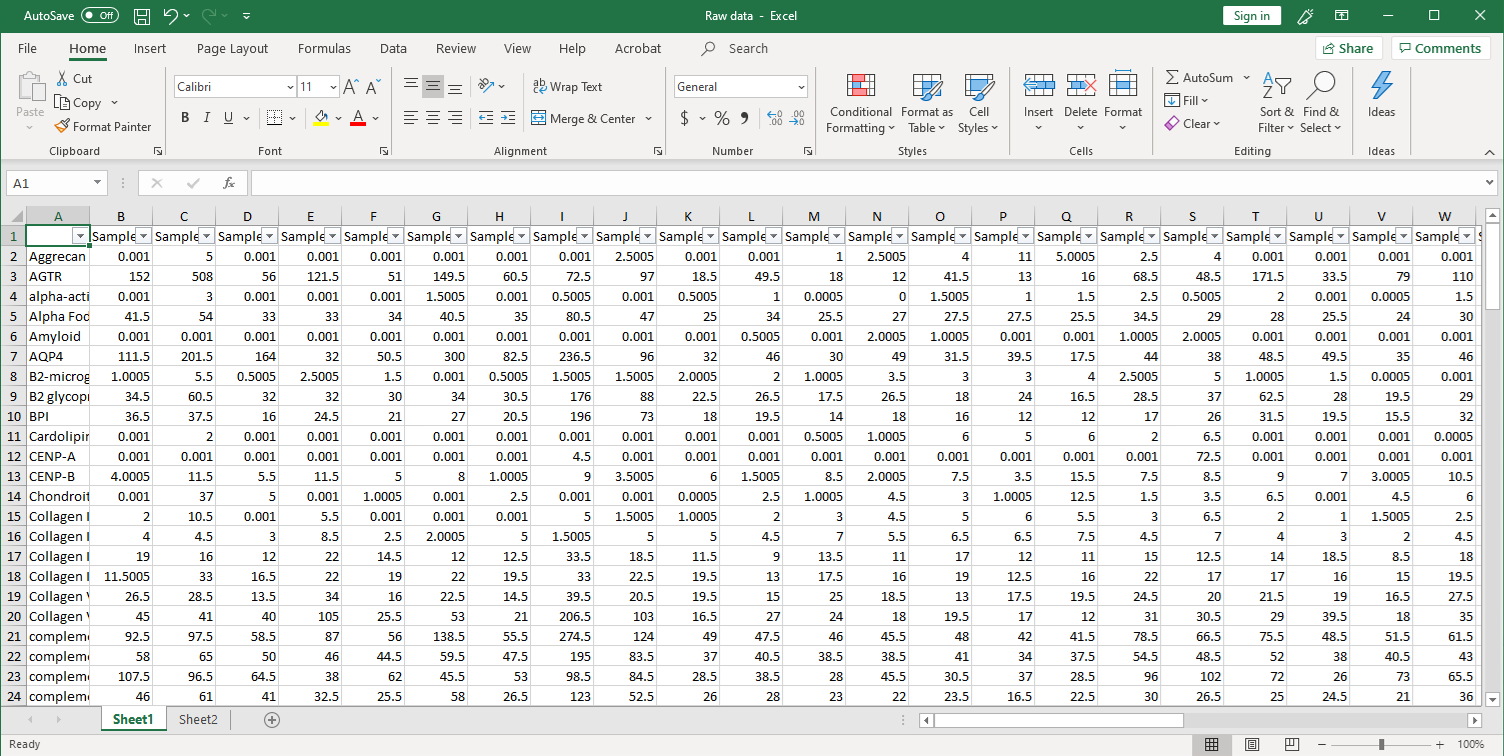
1. Collection of raw data. After scanning, raw signal intensities are collected in an Excel file, as shown below.
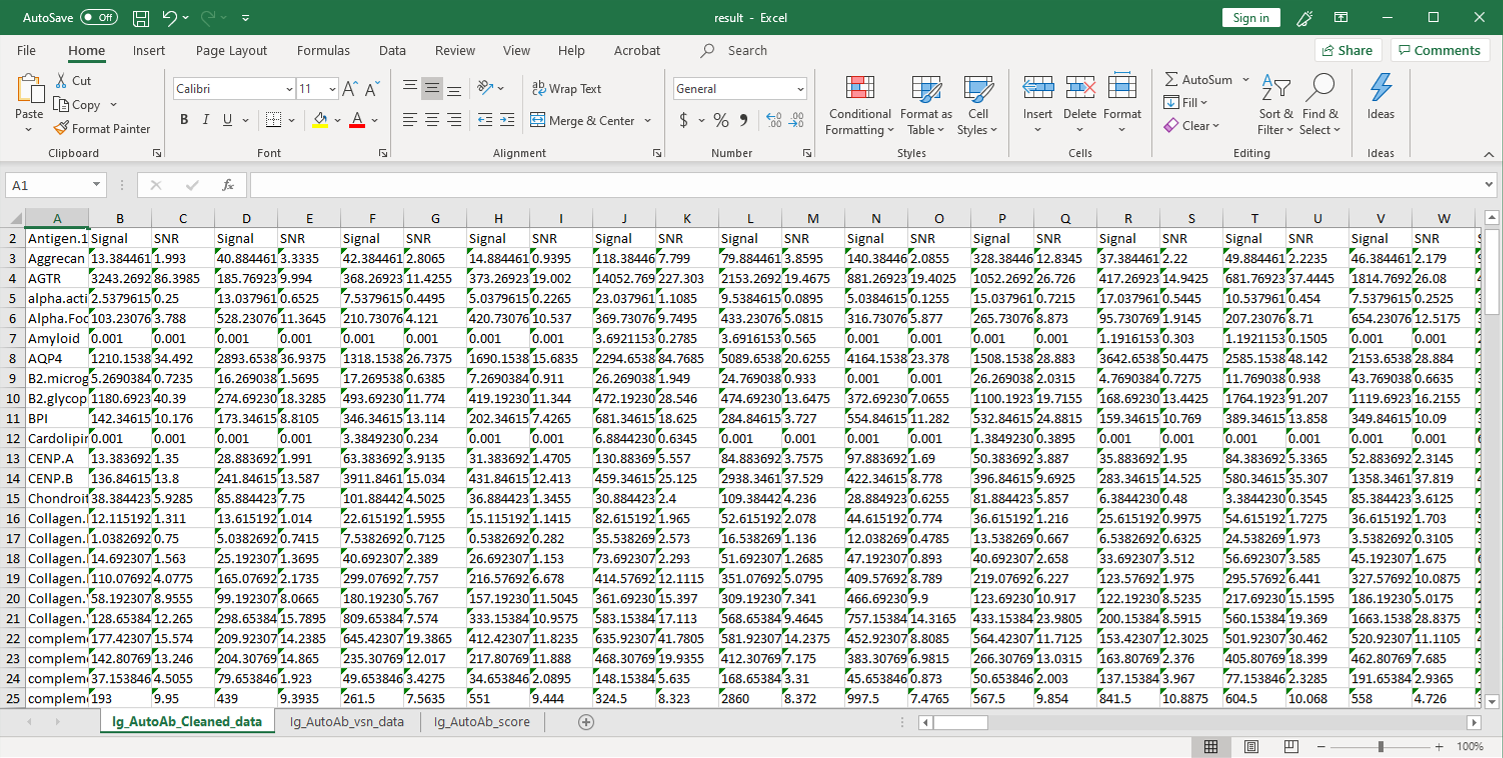
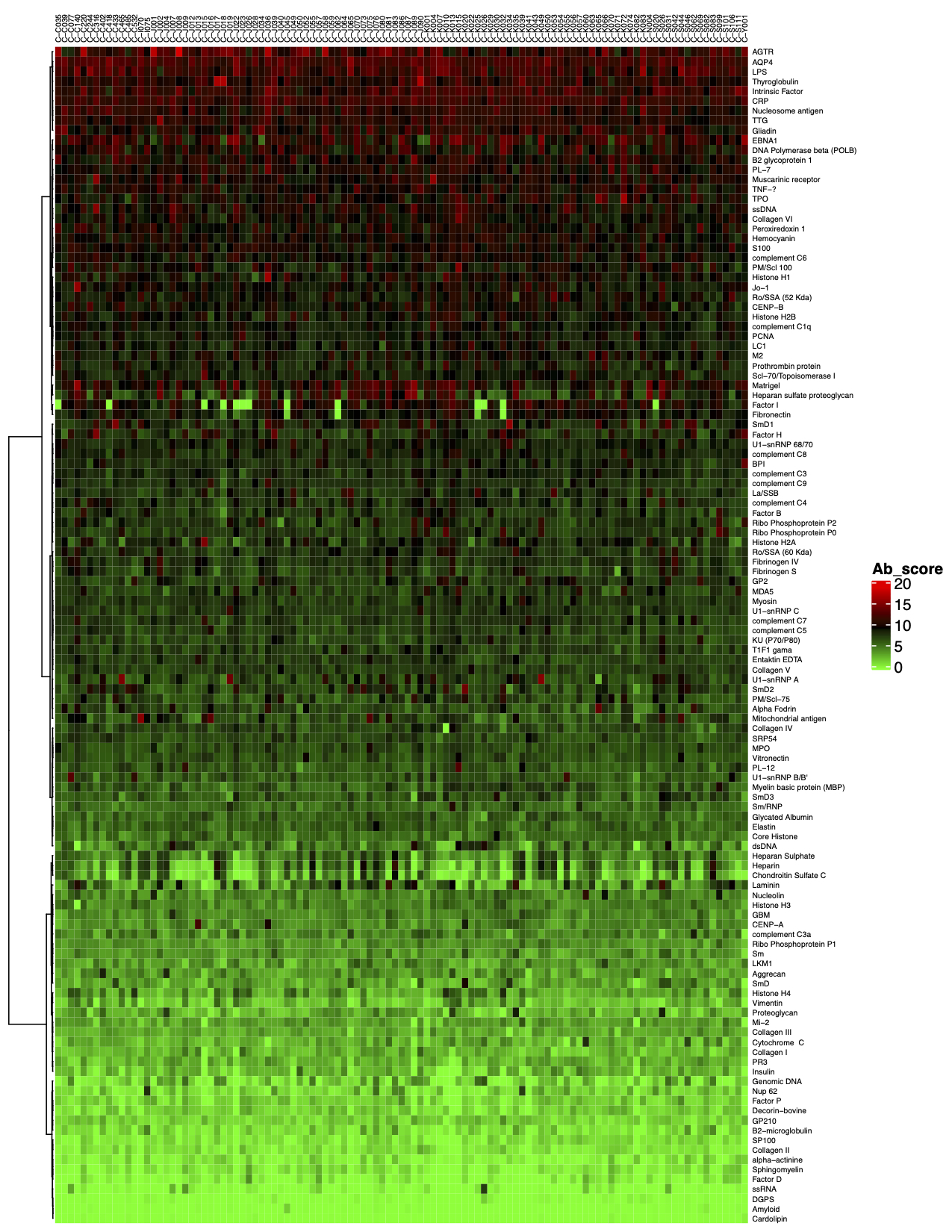
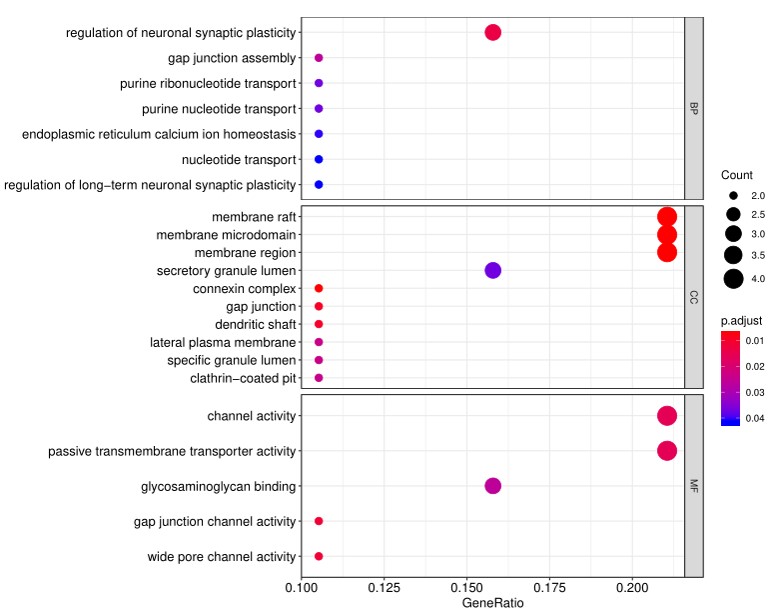
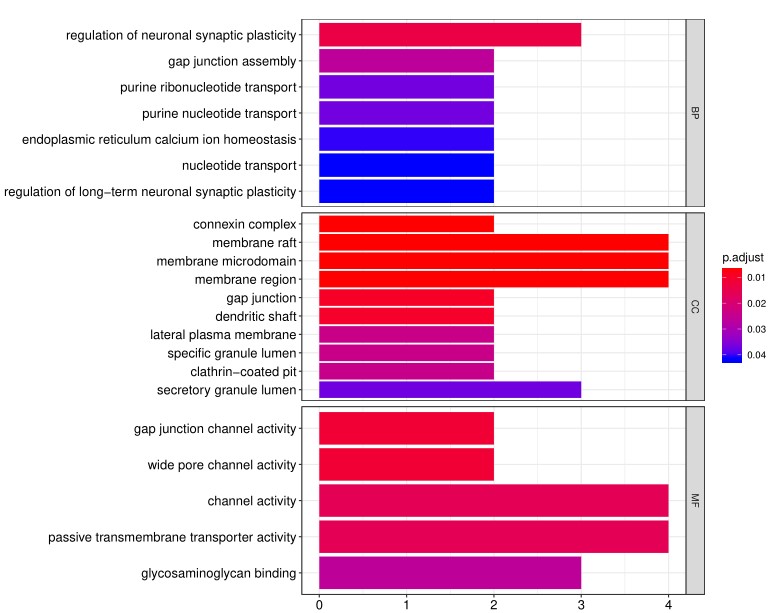
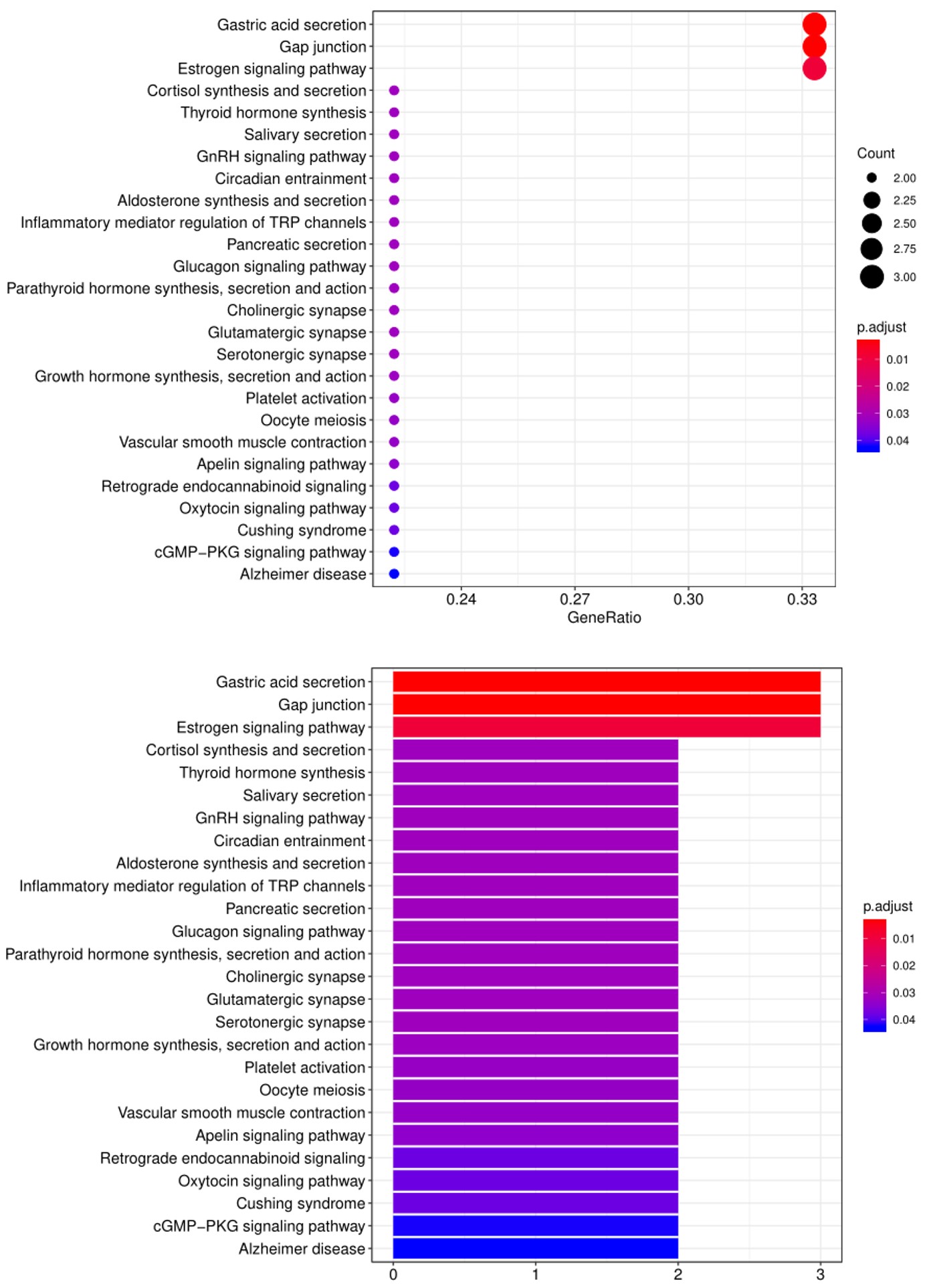
2. Next, the raw data are normalized to controls on the array, and the Net Signal Intensity (NSI) values, as well as the signal-to-noise ratios (SNRs) are tabulated in an Excel file, 3. The final step of the standard analysis package is to determine the “Antibody score” which is the relative enrichment of a given antibody in a sample. The antibody score is determined numerically and displayed in a heat map. A higher score (shifted toward red on the heat map) suggests a stronger antibody-antigen interaction, as shown in the following example: 4. In addition to the standard data analysis package, customers can also choose custom data analysis services. One such service is to classify positive antigens on the array using Gene Ontology (GO) analysis, which classifies genes and proteins based on known biolgical functions. An example of the readout from GO analysis for molecular function is shown below: Further, GO analysis for biological processes is shown in the following example: 5. Another widely-used tool for bioinformatic analysis is KEGG (Kyoto Enclycopedia for Genes and Genomes) Pathway Analysis, which we use to group positive antibody-antigen interactions on the basis of defined biologica pathways, as displayed in the example below:⇒Return to the main OmicsArray™ Antigen Microarray page
Frequently Asked Questions
⇒Return to the main OmicsArray™ Antigen Microarray page
2024
- Li, Q., et al. (2024). Phenotypic and Immunological Characterization of Patients with Activated PI3Kδ Syndrome 1 Presenting with Autoimmunity. J Clin Immunol. 44(4):102.
- Preeti S. Chauhan, et al. (2024). Dietary docosahexaenoic acid supplementation inhibits acute pulmonary transcriptional and autoantibody responses to a single crystalline silica exposure in lupus-prone mice. Front Immunol. 15:1275265.
2023
- Son, K., et al. (2023). Circulating anti-nuclear autoantibodies in COVID-19 survivors predict long COVID symptoms. Eur Respir J. 61(1):2200970.
2022
- Heine, L.K., et al. (2022). Comparative effects of human-equivalent low, moderate, and high dose oral prednisone intake on autoimmunity and glucocorticoid-related toxicity in a murine model of environmental-triggered lupus. Front Immunol. 13:972108.
- Khassawneh, B., et al. (2022). Autoantibody profile in sarcoidosis, analysis from the GRADS sarcoidosis cohort. PLoS One. 17(10):e0274381.
- Ning, W., et al. (2022). Staphylococcus aureus peptidoglycan (PGN) induces pathogenic autoantibody production via autoreactive B cell receptor clonal selection, implications in systemic lupus erythematosus. J Autoimmun. 131:102860.
- Salter, B., et al. (2022). Airway autoantibodies are determinants of asthma severity. Eur Respir J. 60(6):2200442.
- Elias, S., et al. (2022). CXCR4+ Treg cells control serum IgM levels and natural IgM autoantibody production by B-1 cells in the bone marrow. J Exp Med. 219(7):e20220047.
- von Itzstein, M.S., et al. Association between Antibiotic Exposure and Systemic Immune Parameters in Cancer Patients Receiving Checkpoint Inhibitor Therapy. Cancers (Basel). 14:1327.
- Seeley-Fallen MK, Lazzaro M, et al. (2022). Non-Muscle Myosin II Is Essential for the Negative Regulation of B-Cell Receptor Signaling and B-Cell Activation. Front Immunol. 13:842605.
-
Avalos A, Tietsort JT, et al. (2022). Hem-1 regulates protective humoral immunity and limits autoantibody production in a B cell-specific manner. JCI Insight. 7(9):e153597.
-
Labombarde JG, Pillai MR, et al. (2022). Induction of broadly reactive influenza antibodies increases susceptibility to autoimmunity. Cell Rep. 38(10):110482.
- Boustani K, Ghai P, et al. (2022). Autoantibodies are present in the bronchoalveolar lavage but not circulation in patients with fibrotic interstitial lung disease. ERJ Open Res. 8(1):00481-202.
-
Ghosh N, Postow M, et al. (2022). Lower baseline autoantibody levels are associated with immune-related adverse events from immune checkpoint inhibition. J Immunother Cancer. 10(1):e004008.
- Rojas M, Ramírez-Santana C, et al. (2022). New insights into the taxonomy of autoimmune diseases based on polyautoimmunity. J Autoimmun.126:102780.
- Cheng D, Luo Z, et al. (2022). Elevated Cerebrospinal Fluid Anti-CD4 Autoantibody Levels in HIV Associate with Neuroinflammation. Microbiol Spectr. 10(1):e0197521.
2021
- Gonugunta AS, von Itzstein MS, et al. (2021). Humoral and cellular correlates of a novel immune-related adverse event and its treatment. J Immunother Cancer. 9(12):e003585.
- Chiang K, Largent AD, et al. (2021). Cutting Edge: A Threshold of B Cell Costimulatory Signals Is Required for Spontaneous Germinal Center Formation in Autoimmunity. J Immunol. 207(9):2217-2222.
- Hirsiger JR, Tamborrini G, et al. (2021). Chronic inflammation and extracellular matrix-specific autoimmunity following inadvertent periarticular influenza vaccination. J Autoimmun. 124:102714.
- Kiripolsky J, Kasperek EM, et al. (2021). Immune-Intrinsic Myd88 Directs the Production of Antibodies With Specificity for Extracellular Matrix Components in Primary Sjögren’s Syndrome. Front Immunol. 12:692216. doi: 10.3389/fimmu.2021.692216.
- Jacobs HM, Arkatkar T, et al. (2021). TACI haploinsufficiency protects against BAFF-driven humoral autoimmunity in mice. Eur J Immunol. 51(9):2225-2236.
- Cass SP, Dvorkin-Gheva A, et al. (2021). Differential expression of sputum and serum autoantibodies in patients with chronic obstructive pulmonary disease. Am J Physiol Lung Cell Mol Physiol. 320(6):L1169-L1182.
- Pestka JJ, Akbari P, et al. (2021). Omega-3 Polyunsaturated Fatty Acid Intervention Against Established Autoimmunity in a Murine Model of Toxicant-Triggered Lupus. Front Immunol. 12:653464.
- Wilson A, Velasco CA, et al. (2021). Mine-site derived particulate matter exposure exacerbates neurological and pulmonary inflammatory outcomes in an autoimmune mouse model. J Toxicol Environ Health A. 84(12):503-517.
- Kiripolsky J, Kasperek EM, et al. (2021). Tissue-specific activation of Myd88-dependent pathways governs disease severity in primary Sjögren’s syndrome. J Autoimmun. 118:102608.
- Rai P, Janardhan KS, et al. (2021). IRGM1 links mitochondrial quality control to autoimmunity. Nat Immunol. 2021 Mar;22(3):312-32.
- Garimella MG, He C, et al. (2021). The B cell response to both protein and nucleic acid antigens displayed on apoptotic cells are dependent on endosomal pattern recognition receptors. J Autoimmun. 117:102582.
- Luo Z, Alekseyenko AV, et al. (2021). Rigorous Plasma Microbiome Analysis Method Enables Disease Association Discovery in Clinic. Front Microbiol. 11:613268.
2020
- Khan, S., et al. Late-Onset Immunotherapy Toxicity and Delayed Autoantibody Changes: Checkpoint Inhibitor-Induced Raynaud’s-Like Phenomenon. Oncologist. 25: e753.
- Fuchs PS, Lötscher J, et al. (2020). Co-Occurrence of ANCA-Associated Vasculitis and Sjögren’s Syndrome in a Patient With Acromegaly: A Case Report and Retrospective Single-Center Review of Acromegaly Patients. Front Immunol. 11:613130.
- Vo HTM, Duong V, et al. (2020). Autoantibody Profiling in Plasma of Dengue Virus-Infected Individuals. Pathogens. 9(12):1060.
- Turner JA, Stephen-Victor E, et al. (2020). Regulatory T Cell-Derived TGF-β1 Controls Multiple Checkpoints Governing Allergy and Autoimmunity. Immunity. 53(6):1331-1332.
- Raj P, Song R, et al. (2020). Deep sequencing reveals a DAP1 regulatory haplotype that potentiates autoimmunity in systemic lupus erythematosus. Genome Biol. 21(1):281.
- Lammert C, Zhu C, et al. (2020). Exploratory Study of Autoantibody Profiling in Drug-Induced Liver Injury with an Autoimmune Phenotype. Hepatol Commun. 4(11):1651-1663.
- Turner JA, Stephen-Victor E, et al. (2020). Regulatory T Cell-Derived TGF-β1 Controls Multiple Checkpoints Governing Allergy and Autoimmunity. Immunity. 53(6):1202-1214.e6.
- Mayer CT, Nieke JP, et al. (2020). An apoptosis-dependent checkpoint for autoimmunity in memory B and plasma cells. Proc Natl Acad Sci U S A. 117(40):24957-24963.
- Rajasinghe LD, Li QZ, et al. (2020). Omega-3 fatty acid intake suppresses induction of diverse autoantibody repertoire by crystalline silica in lupus-prone mice. J Autoimmunity. 53(7):415-433.
- Perez-Diez A, Wong CS, et al. (2020). Prevalence and pathogenicity of autoantibodies in patients with idiopathic CD4 lymphopenia. J Clin Invest. 130(10):5326-5337.
- Tanaka S, Ise W, et al. (2020). Tet2 and Tet3 in B cells are required to repress CD86 and prevent autoimmunity. Nat Immunol. 21(8): 950-961.
- Mohammed S, Vineetha NS, et al. (2020). Regulatory role of SphK1 in TLR7/9-dependent type I interferon response and autoimmunity. FASEB J. 34(3):4329-4347.
2019
- Chen Y, Yu M, et al. (2019). CXCR5+PD-1+ follicular helper CD8 T cells control B cell tolerance. Nat Commun. 10(1):4415.
- Wang T, Marken J, et al. (2019). High TLR7 Expression Drives the Expansion of CD19+CD24hiCD38hi Transitional B Cells and Autoantibody Production in SLE Patients. Front Immunol. 10:1243.
- Mohammed S, Vineetha NS, et al. (2019). Examination of the role of sphingosine kinase 2 in a murine model of systemic lupus erythematosus. FASEB J. 33(6):7061-7071.
- Ogunrinde E, Zhou Z, et al. (2019). A Link Between Plasma Microbial Translocation, Microbiome, and Autoantibody Development in First-Degree Relatives of Systemic Lupus Erythematosus Patients. Arthritis Rheumatol. 71(11):1858-1868.
- Luo Z, Li M, et al. (2019). Systemic translocation of Staphylococcus drives autoantibody production in HIV disease. Microbiome.7(1):2.
- Mukherjee M, Thomas SR, et al. (2019). Sputum Antineutrophil Cytoplasmic Antibodies in Serum Antineutrophil Cytoplasmic Antibody-Negative Eosinophilic Granulomatosis with Polyangiitis. Am J Respir Crit Care Med. 199(2):158-170.
- Du SW, Arkatkar T, et al. (2019). Functional Characterization of CD11c+ Age-Associated B Cells as Memory B Cells. J Immunol. 203(11):2817-2826.
- Pouzolles M, Machado A, et al. (2019). Intrathymic adeno-associated virus gene transfer rapidly restores thymic function and long-term persistence of gene-corrected T cells. J Allergy Clin Immunol. 145(2):679-697.e5.
2018
- Arkatkar T, Jacobs HM, et al. (2018). TACI deletion protects against progressive murine lupus nephritis induced by BAFF overexpression. Kidney Int. 94(4):728-740.
- Zhang Y, Burberry A, et al. (2018). The C9orf72-interacting protein Smcr8 is a negative regulator of autoimmunity and lysosomal exocytosis. Genes Dev. 32(13-14):929-943.
- Ottens K, Hinman RM, et al. (2018). Foxo3 Promotes Apoptosis of B Cell Receptor-Stimulated Immature B Cells, Thus Limiting the Window for Receptor Editing. J Immunol. 201(3):940-949.
- Palmer VL, Worth AN, et al. (2018). IL10 restrains autoreactive B cells in transgenic mice expressing inactive RAG1. Cell Immunol. 331:110-120.
- Preite S, Cannons JL, et al. (2018). Hyperactivated PI3Kδ promotes self and commensal reactivity at the expense of optimal humoral immunity. Nat Immunol. 19(9):986-1000.
- Chen X, Sun X, et al. (2018). An autoimmune disease variant of IgG1 modulates B cell activation and differentiation. Science. 362(6415):700-705.
Earlier
- Kramer, J.M., et al. Analysis of IgM antibody production and repertoire in a mouse model of Sjögren’s syndrome. J. Leukoc. Biol. 99: 321.
- Packard, T.A., et al. COPD is associated with production of autoantibodies to a broad spectrum of self-antigens, correlative with disease phenotype. Immunol. Res. 55: 48.