Site-specific genome editing at will |
 |
Introduction
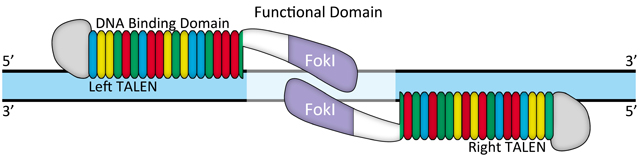
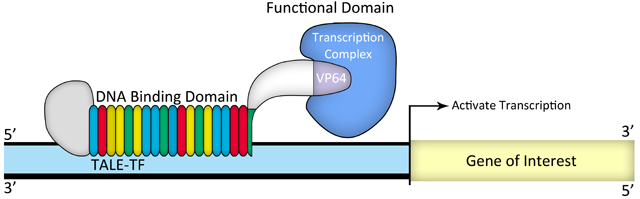
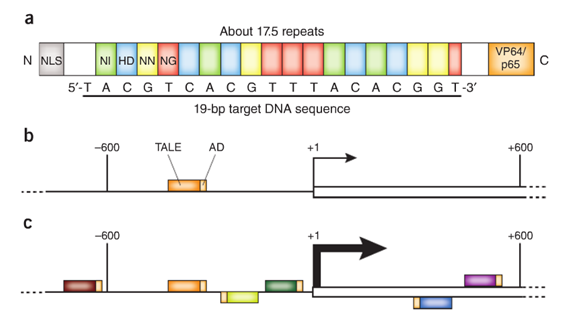
Transcription activator-like (TAL) effectors are DNA binding proteins produced by Xanthomonas bacteria when they infect plants. These proteins can activate the expression of plant genes by recognizing and binding host plant promoter sequences through a central repeat domain consisting of a variable number of ~34 amino acid repeats. The residues at the 12th and 13th positions of each repeat are hyper-variable. There appears to be a simple one-to-one code between these two critical amino acids in each repeat and each DNA base in the target sequence, e.g., NI = A, HD = C, NG = T, and NN = G or A.
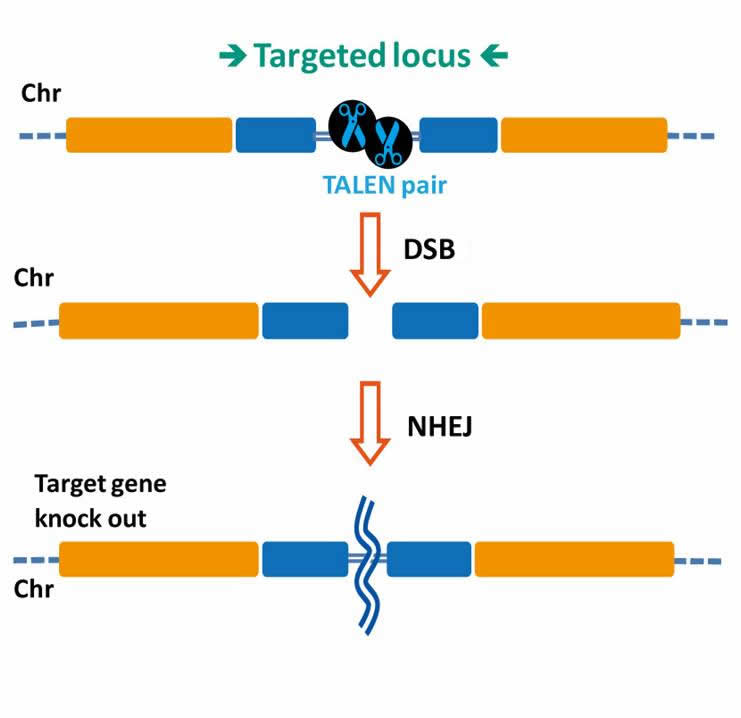
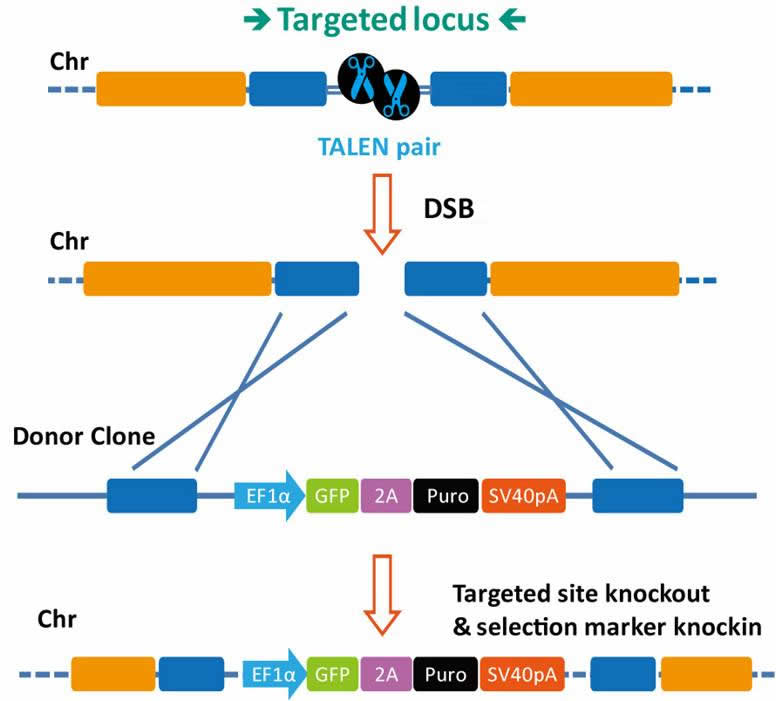
TAL effectors have been utilized to create site-specific gene-editing tools by fusing target sequence-specific TAL effectors to nucleases (TALENs), transcription factors (TALE-TFs), and other functional domains. These fusion proteins can recognize and bind chromosome target sequences specifically to execute their gene editing functions, such as gene knockout, knockin (with donor plasmid), mutagenesis, activation, repression, and more. Unlike zinc fingers’ nucleotide triplet recognition, TAL effector domains recognize single nucleotides, which allows researchers to be able to specifically target whatever sequence they want.
Advantages
- Target any sequence in any cell
- Highly sequence-specific genome editing
- For gene knockout, knockin, mutagenesis, activation, repression and more
- Flexible TAL effector design of binding and functional domains, such as TALEN and TALE-TF
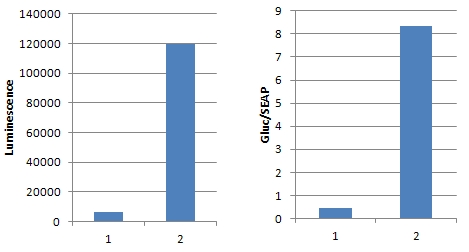
Figure 1. Illustration of TALEN design
Figure 2. Illustration of TALE-TF design
Comparison between TALEN and CRISPR-Cas9 Systems
| Property | TALEN | CRISPR-Cas9 |
|---|---|---|
| Type of recognition | Protein-DNA | RNA-DNA |
| Methylation sensitive | Sensitive | Not sensitive |
| Chromotin structure sensitive | Sensitive | Sensitive |
| Off-target effect | Less observed off-target effects | More potential off-target effects than TALENs and ZFNs |
| Multiplexing | Rarely used | Capable |
References
- Boch, J. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009 326(5959):1509-12
- Moscou, M. et al. A simple cipher governs DNA recognition by TAL effectors. Science. 2009 326(5959):1501
- Christian, M. et al. Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. DOI: 10.1534/genetics.110.120717
- Morbitzera, R. et al. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. www.pnas.org/cgi/doi/10.1073/pnas.1013133107
- Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research, 2011, Vol. 39, No. 12 e82 doi:10.1093/nar/gkr218
- Li, T. et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Research, 2011, Vol. 39, No. 14 6315–6325 doi:10.1093/nar/gkr188
- Zhang, F. et al. Programmable Sequence-Specific Transcriptional Regulation of Mammalian Genome Using Designer TAL Effectors. Nat Biotechnol. 2011 February ; 29(2): 149–153. doi:10.1038/nbt.1775.