|
Ed Davis, Ph.D. |
Genome Editing—making specific changes at targeted genomic sites—is fundamentally important to researchers in biology and medicine (Bogdanove & Voytas, 2011; van der Oost, et al., 2013). Two popular genome editing technologies exploit bacterial systems for adaptive immunity or plant pathogenesis: CRISPR (Clustered, Regularly Interspaced, Short Palindromic Repeats), and TALEN (Transcription Activator-Like Effector Nucleases) and), respectively. Both initiate double-strand breaks (DSBs) at virtually any genomic sequence, and are used for gene knockout, correction of genetic defects, gene tagging, and transgene knockin. GeneCopoeia offers an extensive suite of genome editing tools, including CRISPR sgRNAs and TALEN. We also provide many types of clones and cloning vectors for homology-directed repair (HDR) donors, which are necessary for specific genome editing application. In this Technical Note, we describe the purpose of genome editing donors, how to use them, and how GeneCopoeia can provide you with any kind of donor to suit your genome editing projects.
What is a donor?
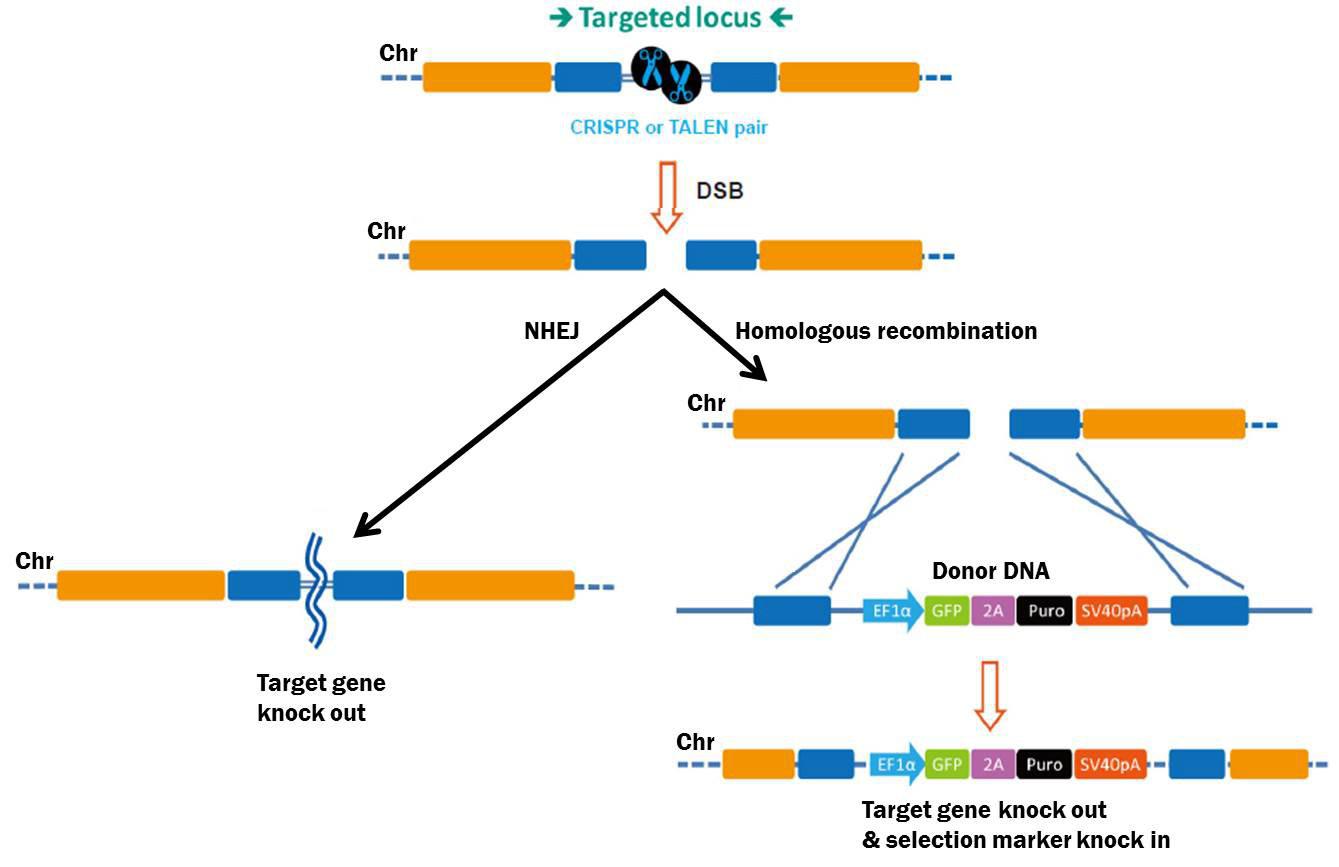
An HDR donor is a DNA molecule that uses HDR (also known as homologous recombination, or HR) to transfer genetic information to a chromosome, and is widely used for knockouts, mutagenesis, gene tagging, promoter swapping, and transgenesis. HDR is initiated by CRISPR- or TALEN-mediated site-specific DSBs. Cells repair DSBs by either of two mechanisms (Figure 1): The first is nonhomologous end joining (NHEJ), which causes small insertions or deletions (“indels”) without a homologous template. This proclivity for indel generation often causes frameshift mutations and is used to knock genes out.
Figure 1. Pathways for repair of DSBs induced by genome editing tools. Left: NHEJ. Right: HDR in the presence of a donor template.
The second major pathway used for DSB repair is HDR. Compared with NHEJ, HDR is relatively error-free Besides sister chromatids, exogenous DNA can also be used as an HDR template. An HDR donor consists of left and right “arms” identical to sequences flanking the DSB (Figure 1). Recombination “copies” information from the donor and “pastes” it to the chromosome. Researchers can place virtually any DNA sequence between the arms—a selectable marker, fluorescent reporter, etc.—and HDR will permit these additional sequences to be copied, or “knocked in” to the chromosome.
Why use an HDR donor?
Most genome editing applications require an HDR donor, except gene knockout. DSBs without donors cause NHEJ-induced frameshift mutations that knock genes out. However, gene knockout can benefit from a donor. First, donors allow precise design of mutations that guarantee knockout. With NHEJ, indels can still preserve the original open reading frame one-third of the time, and sometimes express functional proteins. Also, HDR donors permit drug selection or fluorescence sorting, allowing researchers to avoid time-consuming and labor-intensive screening of large numbers of clones, especially for cultured cell lines with low transfection efficiencies.
So, the decision on whether to use a donor—as well as which type of donor—depends on the application, as described below.
HDR Donor applications
Gene knockout
Gene knockout does not require an HDR donor, and gene knockout HDR donors aren’t useful in transgenic animals, where drug selection or fluorescence sorting is impossible. However, in cell culture, gene knockout donors are valuable. These donors have simple structures. Typically, the left and right homology arms flank the CRISPR or TALEN target sites, such that part of these target sites is deleted and replaced by a cassette carrying a drug selection marker, and, if desired, a fluorescent reporter (Figure 2). Alternatively, donor knockout strategies might involve deleting an entire exon or gene. Since most knockouts require modifying two alleles, two donors, each identical except that they contain two different drug selection markers, might help with clone isolation using double drug selection.
Figure 2. Example knockout HDR donor strategy. HDR donor containing drug selection marker (Puro) and fluorescent reporter (GFP). Drug selection and fluorescence sorting enhance the ability to obtain a knockout. Note: Only one allele is shown for simplicity.
Gene knockout donors can be ordered from GeneCopoeia when searching for gene-specific human and mouse CRISPR or TALEN gene knockout clones. Below, we discuss knock-in donor applications, which are ordered form GeneCopoeia as custom projects
Conditional knockout
Conditional gene knockouts are required when researchers want to knock a gene out, but only at a desired time, in a selected tissue, or under specific physiological conditions. One popular conditional knockout strategy places loxP sites into each intron flanking an exon (a “floxed” allele). The floxed allele is active until expression of Cre recombinase. Cre causes recombination between the loxP sites and deletion of the exon (Figure 3).
Figure 3. Conditional knockout donor. Top: Floxed allele donor with loxP sites flanking an exon. Drug selection and fluorescence sorting allow isolation of clones with the floxed allele. Subsequent treatment with Cre recombinase leads to deletion of the essential exon. Note: Only one allele is shown for simplicity.
As with standard knockout donors, conditional knockout is aided by a drug selection marker or fluorescent reporter (Figure 3). However, for conditional knockouts, the gene must retain normal function before expression of Cre. Therefore, we place the drug selection marker into one of the introns.
Base pair changes
One of the most valuable applications for genome editing is introducing single base pair changes. This is useful for both introducing a mutation into a gene of interest, or repairing a disease mutation.
Two different types of donors for introducing base pair changes into chromosomes are commonly used. The first uses a donor clone with a selectable marker and, if desired, a fluorescent reporter. For this type of donor to be used in cell culture, the cassette carrying the drug resistance and fluorescent reporter genes is placed into a nearby intron (Figure 4).
The second type of mutagenesis donor is a single stranded oligonucleotide (ssODN) with approximately 40-80 nucleotides of homology flanking the site. The chief advantage of ssODN donors is that HDR leads to “scarless” replacement of the endogenous sequence; no other sequences are modified. One example of this approach used an ssODN with CRISPR to correct a mutation causing cataracts in mice (Wu, et al., 2013). However, ssODN donors do not offer the advantage of selection or fluorescence sorting to identify correctly-modified clones in cell culture experiments, and so donor clones with selection and fluorescence markers would be more useful, especially in cell lines that do not transfect well.
Figure 4. Example mutagenesis donor strategy. Top: Donor with a single base pair change. Drug selection and fluorescence sorting allow isolation of clones with the mutated allele. Note: Only one allele is shown for simplicity.
In-frame fusion tagging
The next popular application for genome editing donors is in-frame fusion tagging of a protein. For example, suppose you want to add a C-terminal GFP tag to your endogenous protein. You could use CRISPR or TALEN and a donor to knock in the GFP to the 3’ end of the gene at its native chromosomal locus (Figure 5). As with donors for conditional knockout and mutagenesis, a selection marker is often placed into a nearby intron in order to facilitate isolation of correctly modified clones in cell culture.
Figure 5. Donor-mediated fusion tagging. Donor contains 3’ tag, upstream of the stop codon. Drug selection and fluorescence sorting facilitate clone isolation. Note: Only one allele is shown for simplicity.
Safe Harbor transgene Knockin
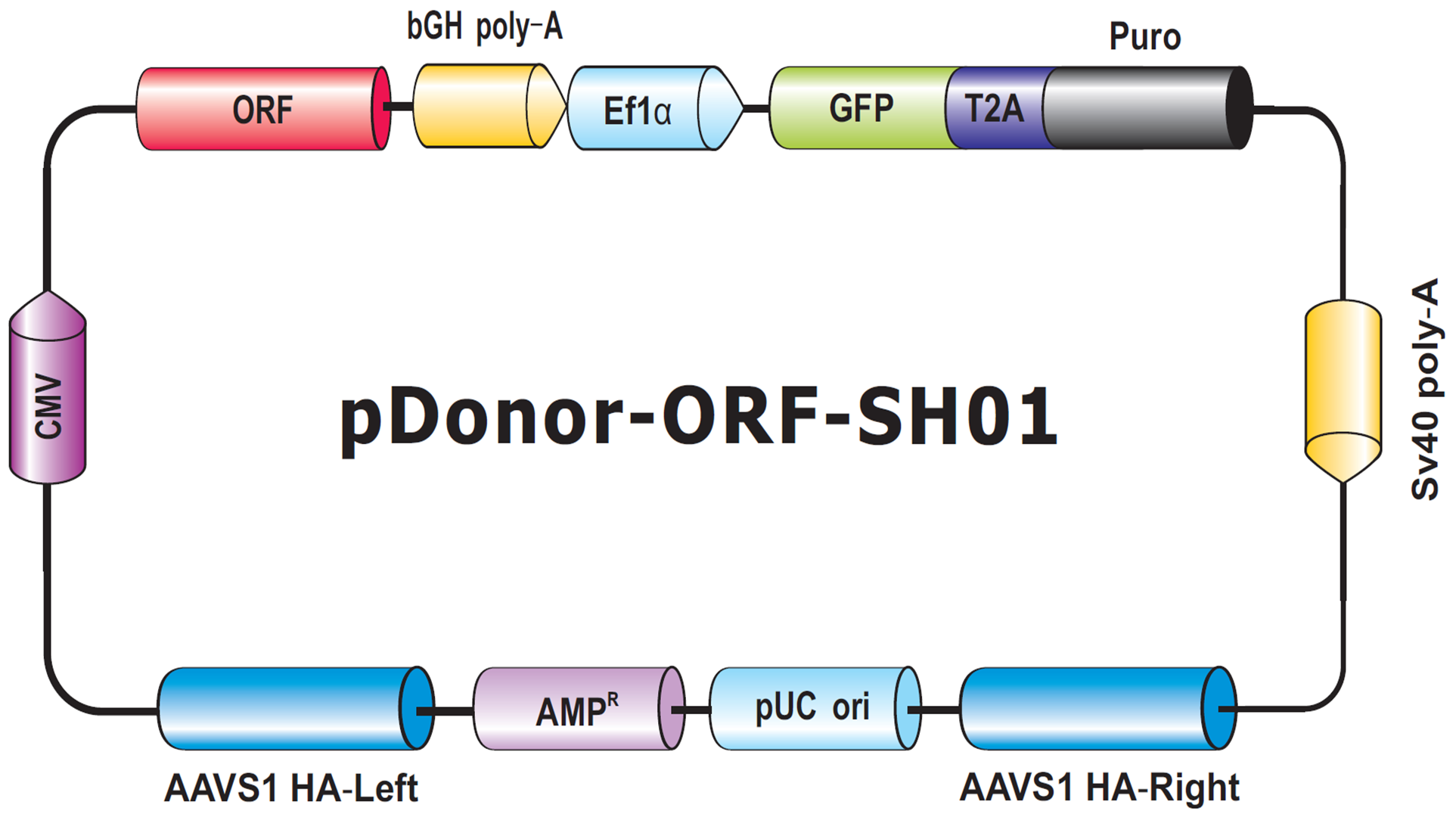
The final type of donor we will discuss in this Technical Note is for transgene knock-in to the human AAVS1 or mouse Rosa26 “Safe Harbor” sites. These sites allow transgene insertion without any deleterious effects on cells or animals, and ensure consistent and stable transgene expression, which often does not occur with randomly integrated transgenes (DeKelver, et al., 2010; Zambrowicz, et al., 1997). Safe Harbor integration has many potential uses, such as integrating a non-human gene into human cells, or rescue of a knockout phenotype with a stably integrated ORF. GeneCopiea provides advanced solutions for Safe Harbor site integration, including kits with validated CRISPR and TALEN reagents, and vectors for do-it-yourself insertion of your transgene of interest between the AAVS1 or Rosa26 homology arms. Further, thanks to GeneCopoeia’s vast collection of human and mouse ORF clones, we offer more than 36,000 Safe Harbor Knockin ORF clones (Figure 6). These clones contain either human or mouse sequence-verified ORFs integrated between the Safe Harbor homology arms.
Figure 6. Genome-CRISPTM Human AAVS1 Safe Harbor Knockin clone backbone. An ORF is placed with a selection cassette between AAVS1 left and right homology arms.
Conclusions
In this Technical Note, we described strategies for HDR donors in genome editing. GeneCopoeia provides many HDR donor solutions, including do-it-yourself donor cloning vectors. However, donor vector design is complex, so we encourage you to use our custom donor cloning services, where our expert Genome Editing team will design, construct, and deliver sequence-verified clones for you. Further, we can construct stable cell lines or transgenic mice containing your CRISPR- or TALEN-mediated modification. For more information about HDR donor vectors and clones visit our website: https://www.genecopoeia.com/product/donor/. You can also call us at 1-866-360-9531, or email inquiry@genecopoeia.com.
References
Bogdanove & Voytas (2011). TAL Effectors: Customizable proteins for DNA targeting. Science 333, 1843.
van der Oost, et al. (2013). New Tool for Genome Surgery. Science 339, 768.
DeKelver, et al. (2010). Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res 20, 1133.
Wu, et al. (2013). Correction of a Genetic Disease in Mouse via Use of CRISPR-Cas9. Cell Stem Cell 13, 659.
Zambrowicz, et al. (1997). Disruption of overlapping transcripts in the ROSA beta-geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. USA 94, 3789.
| Copyright ©2016 GeneCopoeia, Inc. www.genecopoeia.com TNGE5-062216 |