Introduction
For high-throughput profiling of gene expression
The ExProfile™ Gene qPCR Arrays are designed for profiling the expressions of pre-made or customized sets of coding-genes in various tissues or cells. The resulting differential expressions of profiled genes help researchers to identify those that are biologically significant and relevant to their research. In each 96-well plate, there are up to 84 pairs of qPCR primers and 12 wells of controls which are used to monitor the efficiency of the entire experimental process – from reverse transcription to qPCR reaction.
Each pair of primers used in the qPCR arrays has been experimentally validated to yield a single dissociation curve peak and to generate a single amplification of the correct size for the targeted mRNA. A cDNA pool, containing reverse transcript products from total RNA of 10 different tissues, was used as the qPCR validation template.
A universal real-time PCR condition was developed for easy profiling and analysis of the gene expression in a high-throughput fashion. The All-in-One™ First-Strand cDNA Synthesis Kits and qPCR Mix Kits are the recommended and supported RT-PCR reagents for use with the ExProfile™ gene qPCR arrays. These reagents have been optimized to produce high sensitivity, efficiency, and specificity.
Key advantages
Validated mRNA primers
- Each primer pair is designed using a proprietary algorithm and has been experimentally validated
Robust performance
- Sensitive – Detects as low as 4 copies of RNA using ExProfile gene qPCR array and recommended reagents/conditions
- Broad linearity – Simultaneously detects mRNAs at different expression levels
- Reproducible – High reproducibility (R2> 0.99) for inter-array and intra-array replicates
Pre-arranged groups, or customized groups
- Pre-arranged cancer-related groups
- Pre-arranged pathway-related groups
- Customized gene arrays for focused study
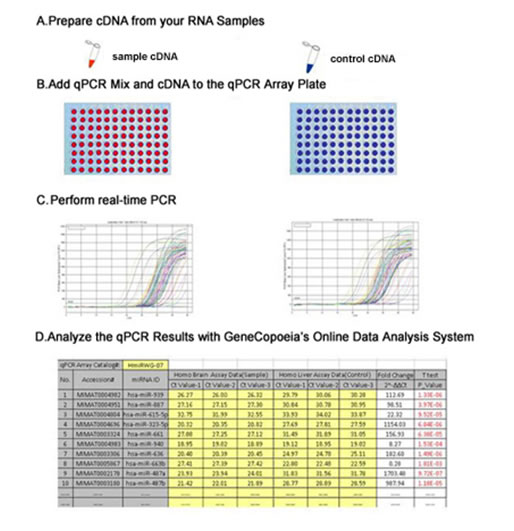
Figure 1. Gene qPCR array experimental work flow.
Performance Data
Linear Range and Sensitivity (spike-in control RNA)
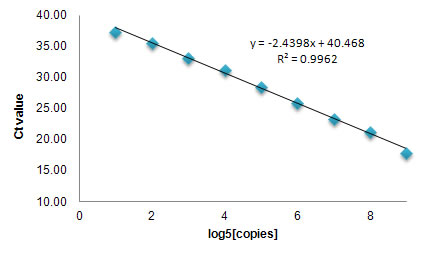 |
Figure 2. Broad linear range and high sensitivity
Mouse total RNA with serially diluted Spike-in control RNA were reverse-transcribed using All-in-One first strand cDNA synthesis kit. The reverse-transcribed cDNA samples were detected using All-in-One qPCR mix and spike-in control specific primers deposited in a 96-well plate. The resulting Ct values were plotted against the log5 of the amounts of spike-in control RNA. The data demonstrated a broad linear dynamic range from 4 to 1.6*106 copies of input RNA as well as high sensitivity. |
Positive calls with a range of total RNA
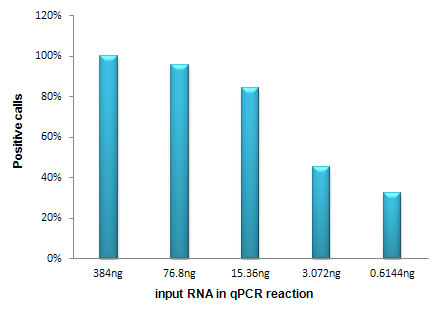 |
Figure 3. High positive calls with as little as 15.36 ng of total RNA
Different amounts of MCF_7 total RNA (1000, 200, 40, 8, 1.6ng) were analyzed with the Human Breast Cancer Gene qPCR Array (PAG-HGBE96-01).The percentage of positive calls (Ct < 35) is plotted against the input amount of total RNA in each qPCR reaction. |
Inter-Array Reproducibility
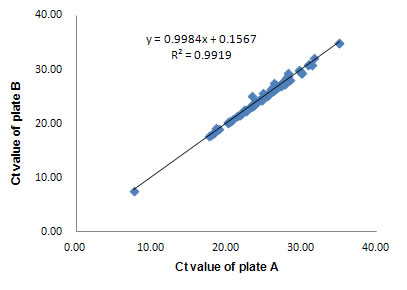 |
Figure 4. High inter-array reproducibility
Two ExProfile™ qPCR gene array replicates (plate A and B) were analyzed using human total RNA (10-tissue mix) on the Bio-Rad iQ5. The Ct values of the replicate plates were plotted against each other. R2 > 0.99 was observed for high inter-array reproducibility. R2 > 0.99 was also observed for intra-array reproducibility (data not shown). |
Catalog Arrays
ExProfile™ Catalog Gene qPCR Arrays
ExProfile™ catalog gene qPCR arrays profile the expression of a group of pre-selected genes, such as pathway-related, cancer-related, disease-relate genes. These genes are carefully chosen for their close correlation to a specific pathway, cancer, or disease, based on a thorough literature search of peer-reviewed publications.
GeneCopoeia offers the following catalog arrays:
ExProfile™ Pathway qPCR arrays
ExProfile™ Cancer Gene qPCR arrays
ExProfile™ Disease and gene group qPCR arrays
User Manual
Custom Arrays
ExProfile™ Custom Gene qPCR Arrays
ExProfile™ Custom Gene qPCR Arrays profile the expression of customer specified genes. Each primer is designed using a proprietary algorithm and has been experimentally validated. Depending on the number of genes and replicates to be analyzed, GeneCopoeia offers 14 layouts to choose from, including 6 types with 96-well plate and 8 types with 384-well plate. Different controls can also be included in the arrays to help monitor the sample quality, RT and PCR reaction efficiencies and replicates reproducibility:
- Genomic DNA control (GDC): detects genomic DNA contamination.
- Spike-in reverse transcription control (RT): monitors the efficiency of the RT reaction.
- Positive PCR control (PCR): verifies the PCR efficiency.
- Housekeeping genes (HK): can be used as endogenous positive controls and for array normalization.
For the best performance, All-in-One™ First Strand cDNA Synthesis Kit and All-in-One™ qPCR Mix are the recommended and supported reagents for use with these arrays.
Key advantages
|
 |
Validated primers
- Each primer is designed using a proprietary algorithm and has been experimentally validated
Flexible array design
- GeneCopoeia offers 14 layouts to choose from, including 6 types with 96-well plate and 8 types with 384-well plate.
Robust performance
- Sensitive – Detects as low as 4 copies of RNA using ExProfile Gene qPCR Array and recommended reagents/conditions
- Broad linearity – Simultaneously detects mRNAs at different expression levels
- Reproducible -High reproducibility (R2> 0.99) for inter-array and intra-array replicates
| Cat. No. |
Product name |
Species |
genes |
Number of plates |
Document |
|
PAG-CS |
Exprofile™ Gene qPCR Arrays Custom Services |
N/A |
N/A |
Variable |
Variable |







