|
Ed Davis, Ph.D. |
Introduction
Genome Editing-the ability to make specific changes at targeted genomic sites-is of fundamental importance to researchers in biology and medicine (Bogdanove & Voytas, 2011; van der Oost, et al.,2013). Two genome editing technologies have emerged recently that exploit bacterial systems for plant pathogenesis or adaptive immunity: TALEN (Transcription Activator-Like Effector Nucleases) and CRISPR (Clustered, Regularly Interspaced, Short Palindromic Repeats), respectively. Both TALEN and CRISPR use endonucleases that initiate double-strand breaks (DSBs) at virtually any genomic target sequence, and are used for many applications, including gene knockout, transgene knock-in, gene tagging, and correction of genetic defects. While both technologies are popular, the decision to choose one technology over the other is not always clear, and GeneCopoeia customers occasionally ask us for advice. In this Technical Note, we compare the advantages and disadvantages of TALEN and CRISPR, with the goal of arming customers with enough information to choose which technology to go with when ordering their reagents from us.
Genome editing
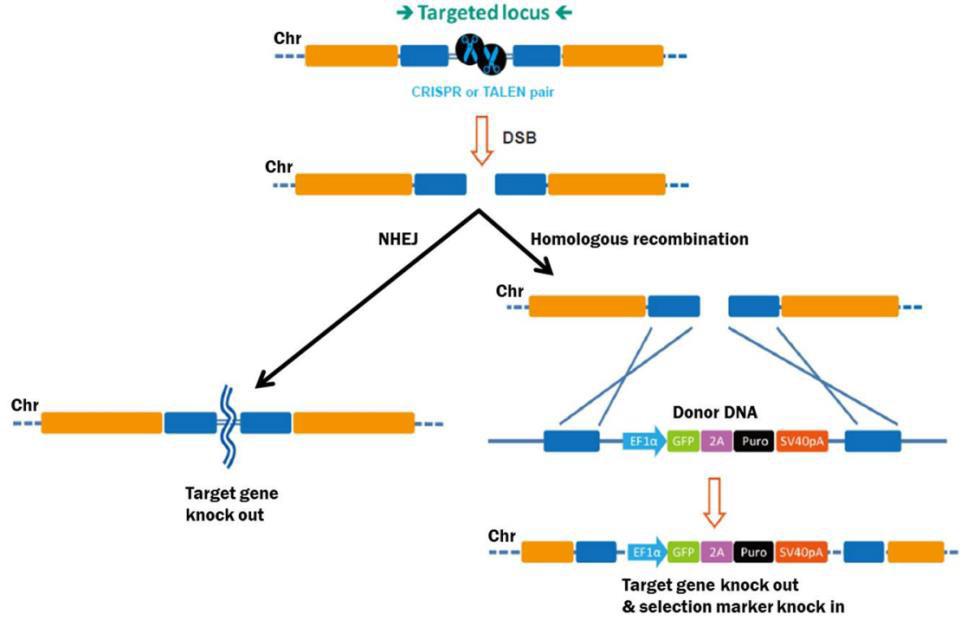
Genome editing starts with efficient DSB generation in the target DNA (Figure 1). DSBs are repaired either by homologous recombination (HR), or, in the absence of a homologous repair template, via non-homologous end joining (NHEJ). NHEJ causes small insertions or deletions as the broken ends are pieced back together. This proclivity for indel generation is exploited as a convenient method for knocking genes out. Both TALEN, which is comprised of a pair of DNA binding proteins fused to the FokI nuclease, and CRISPR, which is a complex between the Cas9 nuclease and a target-specific single guide RNA (sgRNA), can edit DNA through either HR or NHEJ.
Figure 1. Pathways for repair of DSBs induced by genome editing tools. Left: Non-homologous end joining. Right: Homologous recombination in the presence of a donor template.
Comparisons between TALEN and CRISPR
There are four main concerns to think about when trying to decide between TALEN and CRISPR:
1. Specificity
Both TALEN or CRISPR provide high target site specificity, enabling researchers to make precise genetic alterations. CRISPR achieves this specificity through the sgRNA, which is an artificial fusion of two naturally occurring short RNAs (Jinek, et al. 2012). The sgRNA directs the S. pyogenes Cas9 nuclease to a 20 nucleotide target site on the chromosome, which must be immediately followed by an N-G-G trinucleotide known as the Protospacer Adjacent Motif, or PAM (Figure 2). The sgRNA hybridizes with the strand opposite the PAM site, and Cas9 nuclease cuts the DNA. Many recent papers have demonstrated that CRISPR can cause DSB formation at very high frequencies at the intended target site.
However, sgRNAs can tolerate, to varying degrees, up to five mismatches (non-Watson-Crick base pairing) with unwanted target sites (Fu, et al., 2013), and CRISPR has been shown to physically associate with many off-target sites in the genome (Kuscu, et al, 2014). In addition, some studies report very high levels of indel formation at unintended sites (Fu, et al. 2013), although others suggest that the initial estimates of off- target mutagenesis may have been overstated (Li, et al.,2013; Yang, et al., 2013; Wang, et al., 2014).
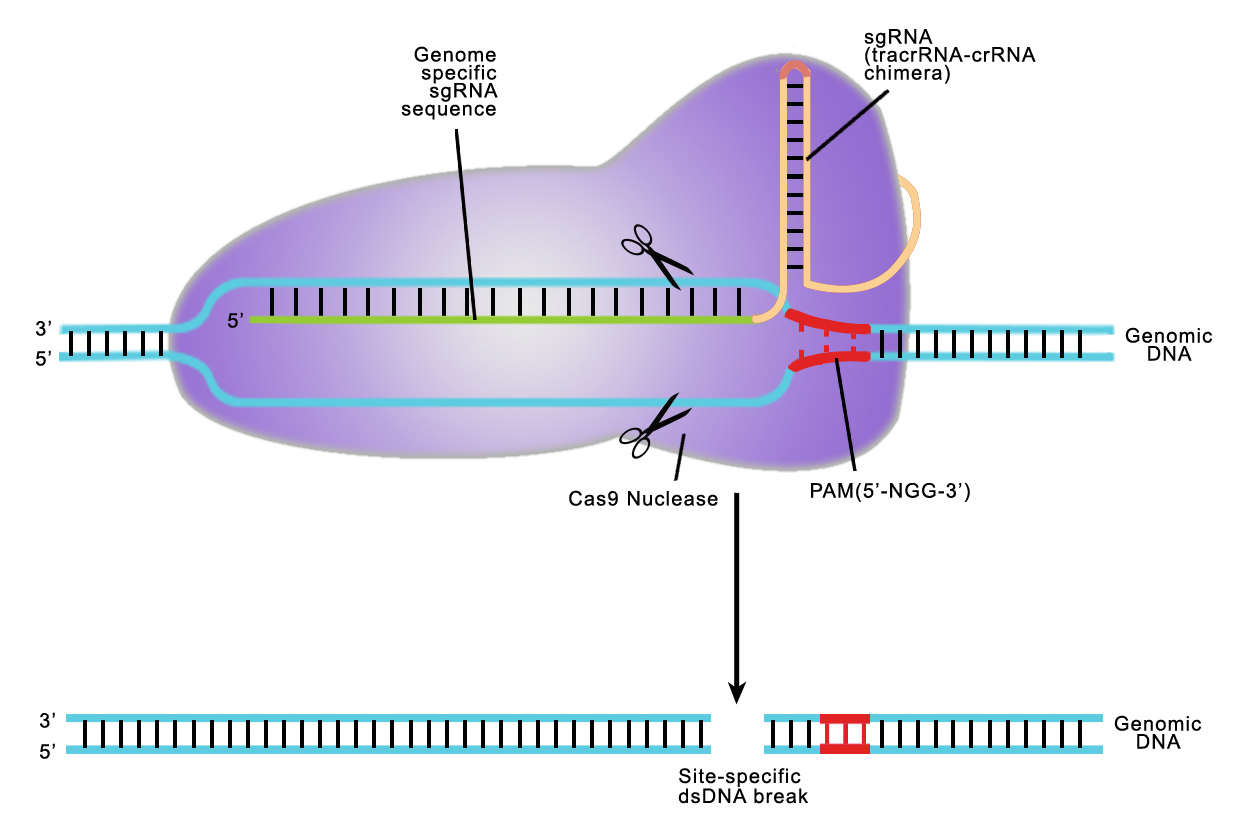
Figure 2. CRISPR-Cas9 target recognition.
Recent developments have significantly improved CRISPR specificity. One strategy employs paired single-strand break (“nickase”) mutants, which require targeting two sgRNAs on opposite strands flanking the target site (Mali, et al., 2013; Ran, et al., 2013). Individual nickases are still recruited to off- target sites, but nicks are less mutagenic than DSBs, so paired nickases dramatically reduce off-targeting. Further, shortening sgRNAs to as few as 17 nucleotides also reduces off-targeting (Fu, et al., 2014). Finally, a fusion between an inactive Cas9 and the FokI endonuclease (RNA-guided FokI nucleases, or “RFNs”; Tsai, et al., 2014), which requires dimerization mediated by offset sgRNA pairs, reduces off- target mutagenesis even more than paired nickases and truncated sgRNAs.
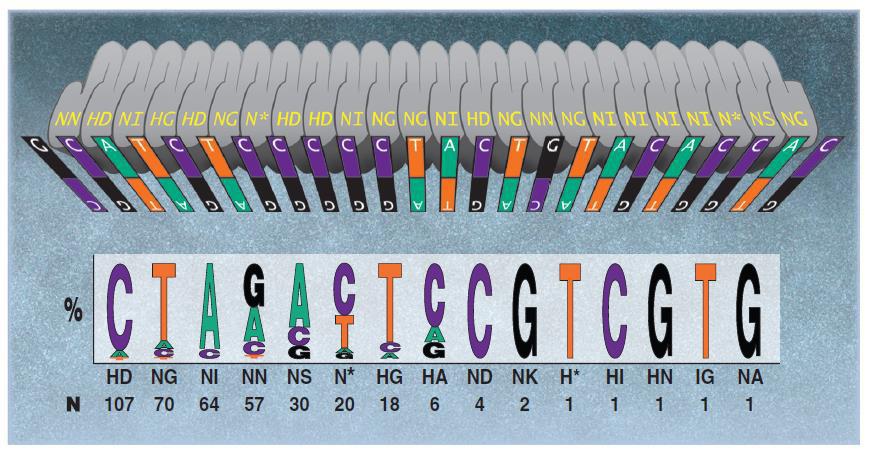
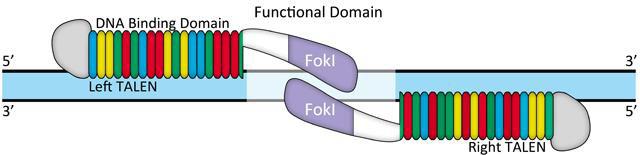
By contrast, off-target activity appears to be less of an issue for TALEN. Typically, TALENs are built with 18 repeats of 34 amino acids. The repeat vary at amino acids 12 and 13, the “Repeat Variable Diresidue”, or RVD. A DNA binding code mediated by the RVD (Figure 3) provides DNA binding specificity. A TALEN pair must bind on opposite sides of the target site, separated by a “spacer” ranging from 14-20 nucleotides (Figure 4). This offset design is necessary because FokI requires dimerization for activity. Therefore, such an extremely long (approximately 36 bp) DNA binding site is expected to be found rarely, if ever, in genomes. There is some degeneracy in the RVD-DNA binding code (Bogdanove and Voytas, 2011), but little evidence of mismatch tolerance or off-target activity has been demonstrated for TALEN. For example, in a recent study, an IPS cell line was edited with a highly active TALEN, and no mutagenic activity was detected at other genome sites homologous to the target site (Park, et al.,2014).
Figure 3. DNA binding code for TALENs. From Bogdanove & Voytas (2011).
Figure 4. Typical TALEN design.
2. Target site selection
As mentioned above, CRISPR sgRNA targets must immediately precede an N-G-G- site. It is usually not difficult to locate GG sites for knockouts, but this constraint sometimes causes problems for other applications. In addition, double nickase and RFN strategies have additional target design constraints. For both, two guide RNAs must be oriented such that the PAM sites are distal from to each other (“tail- to-tail” orientation; Mali, et al. 2013; Ran, et al., 2013; Shen, et al. 2014; Tsai, et al., 2014). Further, the spacing between the sgRNA tails is a critical factor influencing CRISPR design. Paired nickases work best with offset distances between -8 and 30 bp. Optimal spacing between 0 and 20 bp. RFN spacing is even more constrained. 13-18 bp spacing is required, with optimal spacing occurring at 16-17 nucleotides. Combined with the requirement for the PAM sites, the paired CRISPR strategies are sometimes difficult to implement for some applications.
On the other hand, while TALEN design requires offset binding proteins with defined spacing, no other design constraints have yet been described. So in principle, a TALEN pair can be targeted to any site in a genome, which should provide more freedom and flexibility in target site selection than for CRISPR.
3. Efficiency
CRISPR is popular partly because it is capable of modifying chromosomal targets at high frequencies. Rates of indel formation of more than 70% have been reported. The truncated sgRNAs, paired nickases, and RFNs usually have, as expected, lower efficiencies of indel formation.
TALEN is also able to modify chromosomes with high efficiency. In the example cited above (Park, et al.,2014), one TALEN pair had a measured indel formation of 33%. Such efficiency of indel generation is comparable to all CRISPR versions. TALEN, however, is sensitive to cytosine methylation, especially at CpG dinucleotides, which is a common and well-known mechanism for DNA silencing. CpG methylation occurs most often in promoter regions, but it can also occur in protein-coding DNA. Some TALEN pairs provide little to no mutagenesis activity, and this sensitivity to methylation is the likely reason.
4. Ease of design and construction
CRISPR is simple to design and use. For each target site, all that is needed is to program a 20 nucleotide genomic target site into the overall sgRNA. Plasmid construction is straightforward and simple. For editing experiments, the sgRNA is co-expressed with the re-usable Cas9 nuclease. sgRNA design and construction is identical for the paired nickase and RFN approaches, although in these latter cases, the sgRNAs must be chosen and designed to function as a coordinated pair.
1st generation CRISPR has a reputation that target designs are almost always successful. This is not true of the truncated sgRNAs, paired nickases, or RFNs. Some designs are successful, while others are not, even when designed in accordance with published data. Also, CRISPR is not methylation- sensitive.
Even though TALEN construction involves re-engineering a new protein for each target, TALEN design has been streamlined by the availability of modules of repeat combinations that reduce the amount of cloning. Also, as mentioned, TALEN is sensitive to cytosine methylation. One solution is to use the N* RVD for 5-methyl cytosine (Figure 3), but one must know a priori that the target site is methylated. Another workaround for this issue might is to avoid CpG sites. This would add another constraint, of course, but if successful would make TALEN even more attractive as a genome editing tool.
Which should I choose, TALEN or CRISPR?
CRISPR is a newer technology, but TALEN still provides a valuable option, thanks to its relatively unconstrained target site requirements and high degree of specificity. When deciding whether to use TALEN or CRISPR, it is critical to understand the differences between the two systems (Table 1). You should also consider the type of application you are attempting. For example, if you are in a hurry to knock a gene out as part of a quick pilot study, then first-generation CRISPR is a great choice. Need less off-target mutagenesis with relatively unconstrained design? Try TALEN.
Table 1. Comparisons between TALEN and CRISPR.
At GeneCopoeia, we provide products and services for both CRISPR and TALEN. Our genome editing begins with expert design of TALEN and CRISPR vectors in both non-viral and lentiviral formats. In addition, we offer donor plasmid design for HR mediated applications. Plasmid design is followed by construction and delivery of ready-to-use, sequence verified plasmid DNA. Further, we provide more sophisticated genome editing services, such as target site validation, and use of genome editing technology for stable cell line and transgenic mouse line generation. We also offer scientific consulting services, with which you can take advantage of our extensive knowledge and experience in the field. We will help you choose the option most appropriate for your experiments, by informing you of the differences described in this document as well as others, and work with you to devise sound genome editing strategies.
To learn more, visit our genome editing website:
https://www.genecopoeia.com/product/genome-editing/.
References
Bogdanove & Voytas (2011). TAL Effectors: Customizable proteins for DNA targeting. Science 333, 1843.
Fu, et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature Biotechnology 31, 822.
Fu, et al. (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnology 32, 279.
Li, et al. (2013). Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 31, 681.
Jinek, et al. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816.
Kuscu, et al. (2014). Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nature Biotechnology.
Mali, et al. (2013). Cas9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnology 31, 833.
Park, et al. (2014). Targeted inversion and reversion of the blood coagulation factor 8 gene in human iPS cells using TALENs. Proc. Natl. Acad. Scie. U.S.A.
Ran, et al. (2013). Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell 154, 1380.
Shen, et al. (2014). Efficient genome modification by CCRISPR-Cas9 nickase with minimal off-target effects. Nature Methods 11, 399.
Tsai, et al. (2014). Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing van der Oost, et al. (2013). New Tool for Genome Surgery. Science 339, 768.
Wang, et al. (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80.
Yang, et al. (2013). One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas- mediated genome engineering. Cell 154, 1370.
| Copyright ©2014 GeneCopoeia, Inc. www.genecopoeia.com TNGE4-062014 |