|
Ed Davis, Liuqing Qian, Ruiqing lI, Junsheng Zhou, and Jinkuo Zhang |
Introduction
The ability to introduce transgenes into biological systems, such as cultured cells or animals, is of fundamental importance in biology and medicine. Applications for the introduction of such transgenes include, but are not limited to: 1) Overexpression of a protein of interest in order to detect a phenotype; 2) Tagging of a protein with a fluorescent marker for tracking its localization in cells; 3) Rescue of a mutant phenotype with the wild type allele; and 4) Expression of a protein from a particular species, such as mice, in an orthologous organism such as humans. Often, such transgenes are expressed transiently on plasmids. However, in many cases it is desirable to integrate the transgene into the genome so that it is stably passed on to subsequent generations. In this Application Note, we discuss the benefits and drawbacks of such integration, the use of a “Safe Harbor” for transgene integration in mice and humans, and a demonstration of the ability of the GeneCopoeia genome editing tools for Human AAVS1 and Mouse Rosa26 Safe Harbor to integrate transgenes at a “safe” genomic site.
Risks of transgenesis
One classical method for stably integrating transgenes into cells is to transfect or inject linearized DNA and allow random, non-specific integration. While such random integration can be suitable for transgenesis, it carries two major risks: 1) Disruption of a gene leading to lethality or other undesired phenotype; and 2) silencing of the transgene due to local chromatin structure conformation at the integration site. In addition, cells can carry multiple integrants, which can further complicate the interpretation of the effect of the transgene itself. So, the availability of a genomic locus that can accommodate benign insertions that are appropriately expressed is of great utility.
Safe Harbor sites in mice and humans
The first notion of a genomic “safe” site for mammalian transgene insertion came from Phillipe Soriano and colleagues in the late 1990s (Zambrowicz, et al., 1997). They found that one particular strain of mice-named ROSAβgeo26-expressed β galactosidase from a randomly inserted transgene at high levels uniformly in nearly all tissues examined. Subsequent work localized the transgene insertion site to chromosome 6. This locus expresses one coding transcript and two noncoding transcripts, and only the non-coding transcripts are disrupted by the insertion. While pups homozygous for the insertion are born at slightly lower frequency than heterozygous pups, homozygotes appeared to develop normally and were fertile. So, the “Rosa26” locus has since been used as a transgene insertion site that causes no apparent adverse effects on fitness, and permits stable gene expression. By 2010, more than 130 mouse lines containing transgene insertions were generated.
Later, an analogous Safe Harbor site for transgene insertion was described for human cells (DeKelver, et al., 2010). This development grew out of work showing that adeno associated virus (AAV) typically integrates into one genomic locus: PPP1R12C, on chromosome 19. AAV insertions at this locus, also known as AAVS1, do not have any visible phenotype on cells, including primary and immortalized cells, induced pluripotent stem cells (iPSC), and embryonic stem cells. The authors demonstrated that targeted insertions of a transgene at the AAVS1 site caused no apparent harm to any of the cell types examined. In addition, transgenes inserted in order to be expressed from either the native PPP1R12C or an exogenous promoter displayed consistent levels of expression over many cell divisions.
GeneCopoeia genome editing tools for Safe Harbor integration
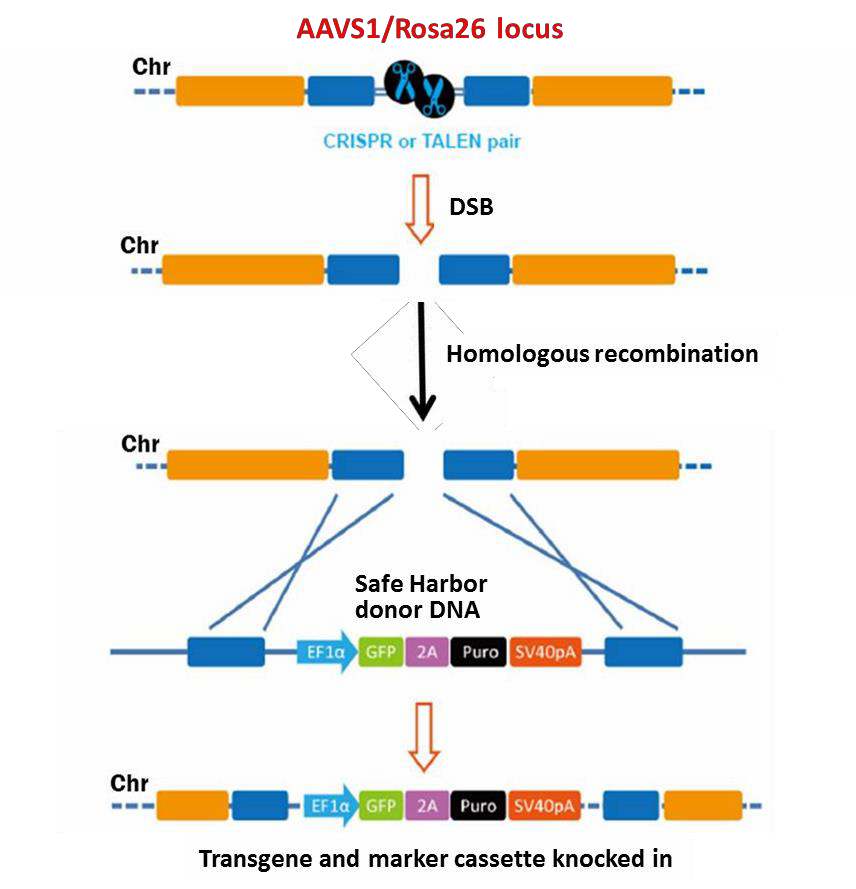
Genome editing is an approach for making specific changes to targeted regions of genomic DNA (for reviews, see Bogdanove & Voytas, 2011; van der Oost, et al., 2013). Two genome editing technologies have emerged recently that exploit bacterial systems for plant pathogenesis or adaptive immunity: TALEN (Transcription Activator-Like Effector Nucleases) and CRISPR (Clustered, Regularly Interspaced, Short Palindromic Repeats), respectively. Both TALEN and CRISPR are chimeric endonucleases that initiate double-strand break (DSB) formation at virtually any target sequence in genomes, leading to mutagenesis. Genome editing has many applications, including gene knock out, gene tagging, and correction of genetic defects, to name a few. In addition, genome editing methodology can be used to introduce transgenes at any locus, including the Rosa26 and AAVS1 Safe Harbor loci. In such a scenario, the TALEN- or CRISPR-induced DSB is repaired by homologous recombination in the presence of a donor
DNA template (Figure 1). Inclusion of desired sequences between recombination “arms” that are homologous to the region flanking the DSB allows the “knock in” of the transgene.
Figure 1. General scheme of the Safe Harbor knock-in strategy using homologous recombination.
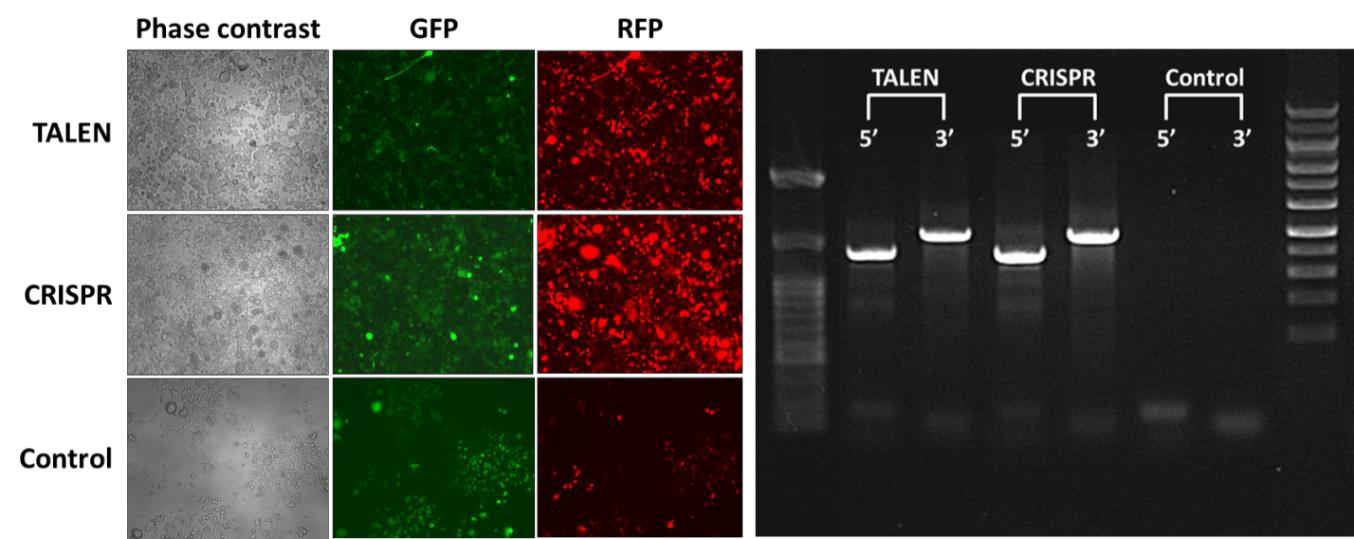
GeneCopoeia has developed tools for genome editing using both TALEN and CRISPR, including reagents for AAVS1 and Rosa26 Safe Harbor. We tested the efficacy of these Safe Harbor tools in cultured cell assays. For Rosa26, we designed three different TALEN nucleases and one CRISPR nuclease that create DSBs at the locus. We next transfected mouse Neuro2a cells separately with each nuclease and a homologous donor construct containing both red fluorescent protein (RFP) and copepod green fluorescent protein (copGFP). As a control, one transfection occurred with only the donor construct. In addition, the transgene contains a marker for puromycin resistance. Treatment of transfected cells with puromycin will select for clones that have integrated the transgene into the genome. If integration occurs, puromycin-resistant cells will be green and red in a fluorescence microscopy assay. Integration of the transgene can occur either randomly or at the Rosa26 site, and so PCR using primers spanning the intended junction site is employed to screen for targeted homologous recombination.
The results of the transfection are shown in Figure 2. Fluorescence assays indicate that both TALEN and CRISPR induced higher levels of integration of the transgene over the cells containing only the donor plasmid. Further, when we isolated genomic DNA from harvested cells and performed PCR with primers spanning the junction of the transgene with the Rosa26 site, we found that only those cells that had been transfected with either TALEN or CRISPR were positive for this assay (Figure 2, right). It should be noted that we did not assay the cells for random integration of the transgene. Nonetheless, the results clearly show that GeneCopoeia TALEN and CRISPR reagents are capable of facilitating high levels of integration at the mouse safe harbor locus.
Figure 2. TALEN- and CRISPR-mediated transgene integration at Rosa26. Left: Neuro2a cells co- transfected with either a TALEN or CRISPR and a donor plasmid, or donor plasmid alone (Control). Green and red fluorescence was assessed after 12 days of puromycin selection. Right: Results of junction PCR for TALEN, CRISPR, and Control cells. Two additional TALENs also mediated Rosa26 site-specific integration (data not shown).
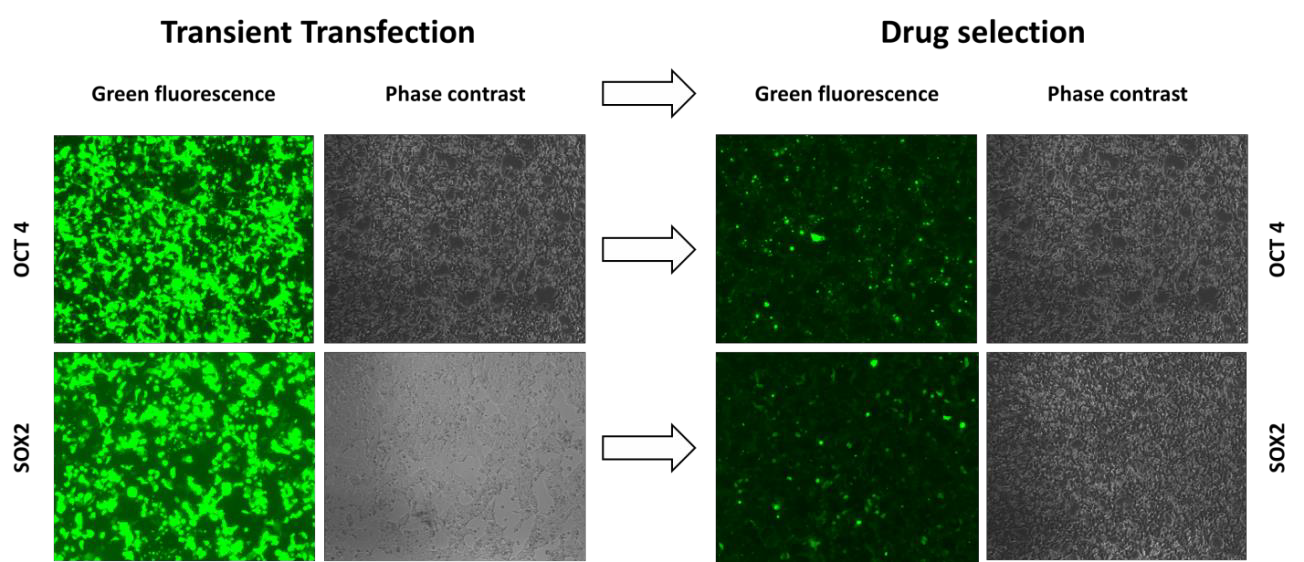
GeneCopoeia also has tools for site-specific transgene integration at the human AAVS1 locus on chromosome 19. In an assay similar to that used for the Rosa26 experiment, we transfected HEK293T cells with AAVS1 TALEN and donor plasmids carrying transgenes with either the SOX2 or OCT4 genes. The transgene also contained the puromycin resistance gene and GFP as a fluorescent marker. We were able to easily identify GFP-expressing cells after 14 days of puromycin selection (Figure 3). Although we again did not rule out random transgene integration in these cells, PCR analysis using primers spanning the 5’ and 3’ junctions showed that cells transfected with the TALEN and donor constructs contained integration of the transgenes at the AAVS1 site (data not shown).
Figure 3. GeneCopoeia TALEN for Human AAVS1 Safe Harbor stimulates site-specific integration of two different transgenes. GFP and phase contrast images were taken both before (left, Transient Transfection) and after 14 days of culturing in the presence of 1μg/ml puromycin (Drug selection, right).
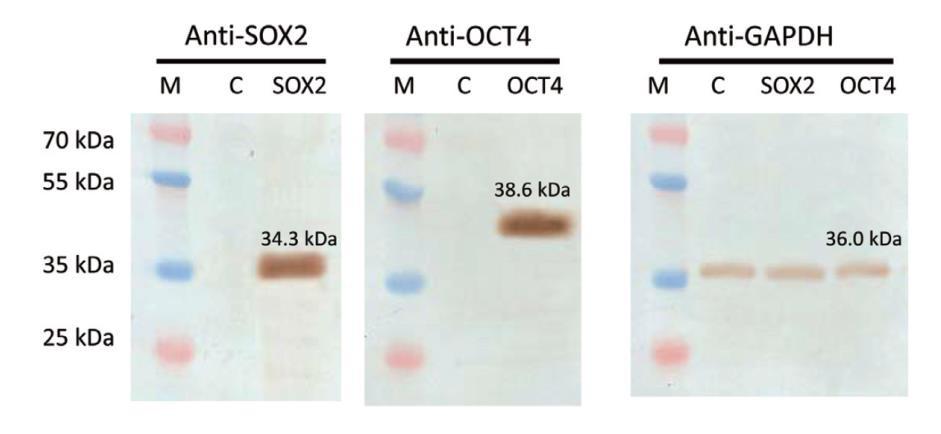
In addition, we analyzed these cells for SOX2 and OCT4 expression by immunoblotting (Figure 4). The results demonstrated that our SOX2- and OCT4-containing transgenes expressed the correct proteins at levels far above background.
Figure 4. Western blot analysis of cells containing SOX2 and OCT4 ORF transgenes integrated at the human AAVS1 Safe Harbor site. Membranes were probed with protein-specific antibodies. M: Marker. C: Untransfected cells. Anti-GAPDH was used as a loading control. Endogenous Sox2 and Oct4 protein levels were below the detection limit in this assay.
GeneCopoeia Human AAVS1 Safe Harbor Knock in Clones
As an enhancement to our Human and mouse Safe Harbor knock in tools, GeneCopoeia offers Safe Harbor Knock in ORF clones for the human AAVS1 site. GeneCopoeia is the original manufacturer of transcriptome-wide, ready-to-express mammalian ORF clones. For the human Safe Harbor technology, we have extended developed a set of more than 18,000 human ORFs inserted into our donor clone designed for knock in at the AAVS1 site (Figure 5).
Figure 5. GeneCopoeia donor vector for Human AAVS1 Safe Harbor Knock in ORF clones. The “ORF” module (in red) represents any of the more than 18,000 human ORF sequences available for Safe Harbor Knock in.
Summary
The discovery of Safe Harbor sites for transgene integration in mice and humans has been a boon for researchers and industries needing consistent protein expression without physiological consequences. An even bigger boost to the field was provided by revolutionary new developments in genome editing technology. For more information about GeneCopoeia’s leading-edge tools for Safe Harbor integration, and other genome editing solutions, please visit our website: www.genecopoeia.com.
References
Bogdanove & Voytas (2011). TAL Effectors: Customizable Proteins for DNA Targeting. Science 333, 1843.
DeKelver, et al. (2010). Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res 20, 1133.
van der Oost, et al. (2013). New Tool for Genome Surgery. Science 339, 768.
Zambrowicz, et al. (1997). Disruption of overlapping transcripts in the ROSA beta-geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. USA 94, 3789.
| Copyright ©2014 GeneCopoeia, Inc. www.genecopoeia.com ANGE1-040414 |