| Ed Davis, Ph.D. |
Abstract
Genome Editing-the ability to make specific changes at targeted genomic sites in complex organisms-is of fundamental importance in biology and medicine (Bogdanove & Voytas, 2011; van der Oost, et al.,2013). Recently, the CRISPR (Clustered, Regularly Interspaced, Short Palindromic Repeats)-Cas (CRISPR- associated) system has become popular for applications such as gene knockouts, making precise, defined base changes, and for transgenesis, to name a few. The ease of design, high efficiency, and relatively low cost of CRISPR-Cas offers promise for use of this tool for correcting mutations that cause genetic diseases, and to replace older methods that cause undesired consequences of random transgene integration. However, CRISPR-Cas itself has some propensity for causing off-target mutagenesis. Despite recent improvements in the technology, some researchers believe that CRISPR-Cas has a relatively low degree of specificity. In this Technical Note, we discuss the mechanism of CRISPR-Cas in its application for genome editing and how it affects specificity, reports in the literature discussing CRISPR’s potential for off-target mutagenesis, approaches for increasing the target specificity of CRISPR- Cas, and GeneCopoeia solutions for improving CRISPR-Cas specificity.
CRISPR-Cas target recognition
The most widely-used version of CRISPR-Cas in genome editing applications employs two components:
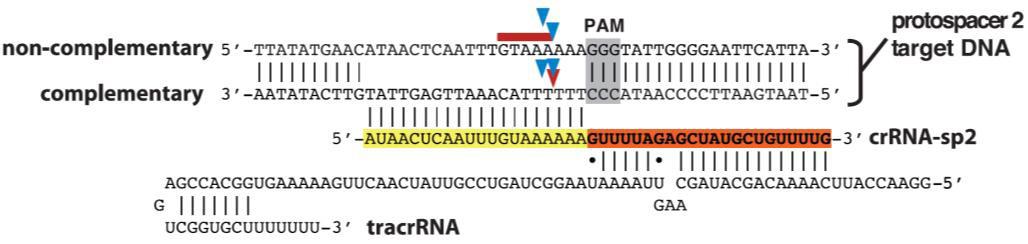
1) The Cas9 nuclease from Streptococcus pyogenes; and 2) A single guide RNA (sgRNA). The sgRNA is a chimera of two RNA molecules of the CRISPR target recognition system, as first described by Jinek, et al. (2012): The CRISPR RNA (crRNA) and the transactivating CRISPR RNA (tracrRNA; Figure 1). An sgRNA contains a 20 nucleotide variable region that provides target site specificity. In addition, the S. pyogenes Cas9 requires a 3 nt N-G-G sequence, known as a “PAM” (Protospacer Adjacent Motif), immediately following the specificity sequence, for DNA binding. The PAM must be on the same strand as the guide RNA sequence, but not part of it.
Figure 1. S. pyogenes Cas9 nuclease requires the presence of two RNAs, the target-specific crRNA, and the structural tracrRNA, for binding and cleavage. The target site is highlighted in yellow. The PAM site is highlighted in gray. Arrows indicate potential nuclease cutting sites. The red arrow is the most frequent cutting position. These RNAs can be fused into one sgRNA for genome editing. From Jinek, et al. (2012).
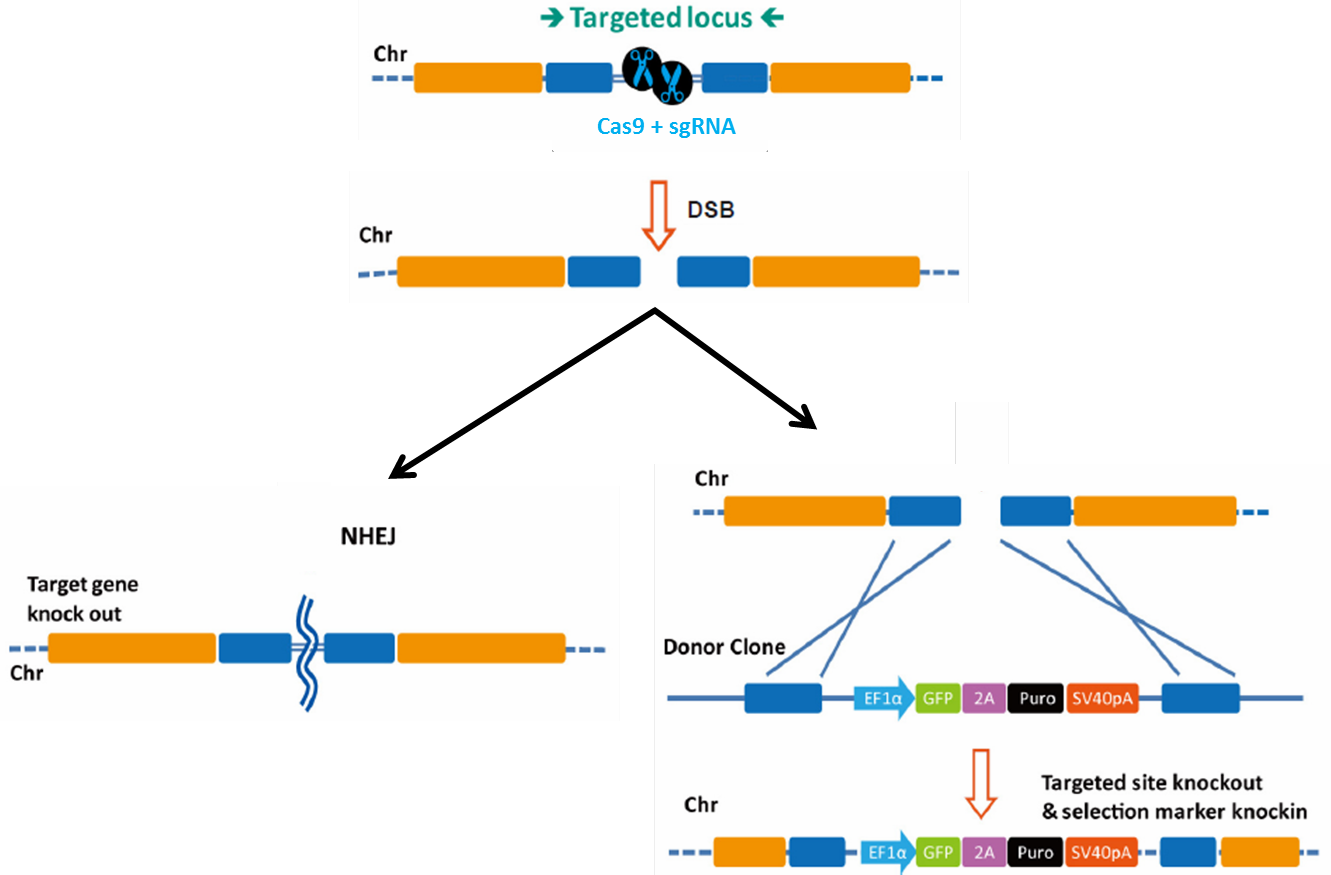
When the sgRNA forms a complex with Cas9, the complex locates the target and PAM sequence, unwinds the DNA duplex, and the 20nt guide RNA anneals to the complementary sequence on the opposite strand. This enables the Cas9 nuclease to create a double-strand break (DSB; Figure 1). DSBs must be repaired immediately or the cell will die. The cell’s preferred pathway of DSB repair is the relatively error-free mechanism of homologous recombination (HR), but this is almost never available in mitotically dividing cells because it must occur during anaphase, after chromosomes have duplicated and sister chromatids have paired. HR also usually doesn’t occur in non-dividing cells. Instead, in the absence of homology, DSB repair occurs via non-homologous end joining (NHEJ). NHEJ is extremely error-prone, leading to the formation of small insertions or deletions (indels) as the broken chromosome ends are pieced back together. This proclivity for indel generation has been exploited by researchers using CRISPR-Cas as a simple, efficient method for knocking genes out.
Figure 1. Pathways for repair of DSBs induced by genome editing tools. Left: Non-homologous end joining. Right: Homologous recombination in the presence of a donor template.
Specificity of sgRNA binding
A given CRISPR-Cas9 target sequence is 20 nt long, so, if binding of the RNA to the chromosomal target were based strictly on sequence, these targets should be unique in eukaryotic genomes. In combination with the, PAM, it should be extremely unlikely that CRISPR-Cas would make another cut.
But there’s more to CRISPR specificity than just sequence composition. Yanfang Fu and colleagues reported that sgRNAs can guide the Cas9 nuclease to three different targets in a chromosomally- integrated GFP reporter gene, despite the presence of one or more mismatches (Fu, et al., 2013).
Dispersed as well as adjacent multiple mismatches between the guide RNAs and the reporter target could be tolerated as well. Mismatches in the 10 nucleotides of the guide RNA most proximal to the PAM site were usually-though not always-tolerated to a greater degree than those in the PAM-distal 10 nucleotides, and the distribution tolerance of mismatches varied depending on the target.
In addition, Fu, et al., generated guide RNAs for three endogenous human genes and predicted, based on the reporter assay results, additional off-target sites in the genome for each. Nearly all off-target sites contained multiple mismatches with each target RNA. Using the T7 Endonuclease I assay to detect the presence of indels, Fu, et al. found that these predicted off-target sites frequently sustained indels, often as much or more than the intended target sites. Up to 5 mismatches between the guide RNA and the chromosome could still allow detectable indel formation. The degree of mismatch tolerance varied depending on the guide RNA sequence, the chromosomal targets analyzed, and the cell lines used.
Recent analysis of CRSIPR-Cas off-target mutagenesis
Early tests of CRISPR-Cas specificity such as those by Fu, et al. (2013) cast doubt on the viability of using this technology for applications requiring high specificity, such as gene therapy. Newer studies, though, have reported better specificity for CRISPR-Cas. For example, Eric Lander’s lab at MIT used pools of lentiviral based sgRNAs for large-scale mutagenesis screens in human cells. Screens for resistance to DNA damaging agents demonstrated that mutagenesis of genes in the mismatch repair pathway occurred at much greater frequencies by correctly-targeted sgRNAs than by incorrectly-targeted guides (Wang, et al., 2014). In addition, an sgRNA that was used to successfully restore wild-type function to the Crygc gene and cure cataracts in mice had no effect in 10 of 12 transgenic animals at 10 predicted off-target sites, and only 1 site was mutated in each of the remaining 2 mice (Wu, et al., 2013). These and other studies (Li, et al., 2013; Yang, et al., 2013), combined with improved software tools for predicting sgRNAs with high potential specificity (Ran, et al., 2013a; Xiao, et al., 2014), suggest that lingering worries that CRISPR-Cas9 has a high propensity for off-target mutagenesis might be overstated.
GeneCopoeia solution to CRISPR-Cas off-target mutagenesis
GeneCopoeia uses sophisticated bioinformatics tools to predict potential genomic sites for Cas9 sgRNA off-target binding. Whether the application is knockout, knockin, introduction of base changes, etc., we design CRISPR sgRNAs that are most likely to function properly, while at the same time discarding those guides with a high likelihood for causing off-target mutagenesis.
Cas9 variants for increased specificity
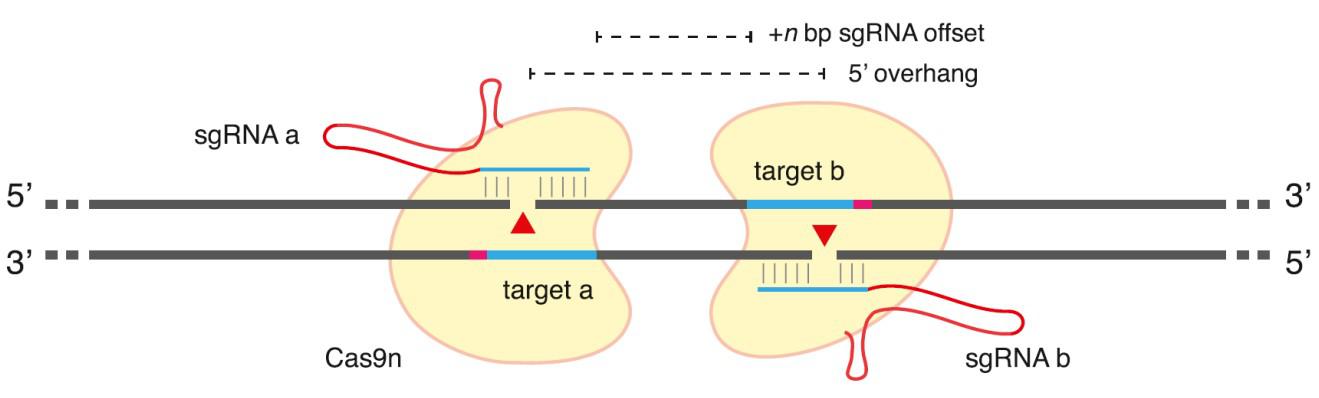
In addition to improved guide RNA design, mutant forms of Cas9 have emerged to further decrease CRISPR-mediated off-target mutagenesis. Wild-type Cas9 has two nuclease domains, each of which cut one DNA strand. Mutations in either domain alone convert Cas9 into a “nickase”, which makes only a single-strand break. While the nickase mutants do not change sgRNA binding specificity and can still be delivered to off-target sites, single nicks have much lower potential for causing mutagenic NHEJ than DSBs. However, when two guide RNAs are designed such that each binds the opposite strand of the chromosome, the result is a staggered-cut DSB that can be repaired by NHEJ or HR (Figure 2). Two groups-one led by George Church at Harvard and the other by Feng Zhang at MIT, showed that this “paired nickase” strategy efficiently led to DNA repair by both NHEJ and HR (Ran, et al., 2013b; Mali, et al., 2013). Zhang’s group further demonstrated that the paired nickase strategy dramatically reducedthe frequency of off-target mutagenesis from 50-fold to more than 1,500 fold for sgRNAs targeted to three distinct genes.
Figure 2. General scheme of Cas9 double-nickase strategy. From Ran, et al. (2013b).
Double nickase design is not easy, though. Guide RNAs oriented with the PAM sites most distal from one another (a “tail-to-tail” orientation; Shen, et al., 2014) lead to indel formation at much higher frequencies than those oriented with the PAM sites proximal to one another (“head-to-head”; Ran, et al., 2013b; Mali, et al., 2013). In addition, the spacing between the ends of each guide sequence (the offset distance) is important. The most efficient nickase pairs generally occur with offset distances between 0 and 20 base pairs. So while the double nickase approach works well to efficiently create DSBs with few consequences resulting from off-target cleavage, the design constraints limit the potential range of suitable sites for some applications.
Further, Keith Joung’s lab published two papers describing two new potential Cas9 alternative strategies that also might elevate the level of CRISPR-Cas specificity. One of the papers showed that sgRNA sequences with as few as 17 nucleotides were usually as effective at creating indels as the standard length of 20 nucleotides (Fu, et al., 2014). Off-target mutagenesis caused by these truncated sgRNAs targeted to four genes was reduced by up to 5,000 fold. In the other paper (Tsai, et al., 2014) Joung’s lab fused a Cas9 mutant with no nuclease activity (Cas9n) to the nuclease domain of FokI. FokI requires dimerization to cut, so, in a fashion similar to the double nickase strategy, a Cas9n-FokI pair was shown to dramatically reduce off-targeting. However, it remains to be seen whether the approaches of the truncated sgRNAs, which don’t always function at high efficiency, or the Cas9-FokI fusion, which has spatial design constraints, will be effective in general.
GeneCopoeia solutions for increased Cas9 specificity
GeneCopoeia scientists are experts at Cas9 D10A double nickase design strategy. We diligently predict the paired guide RNAs that have the highest probability of efficient cutting, based on standards reported in the literature, as well as our own proprietary requirements. Our double nickases are designed to function either as a pair with the Cas9 D10A nickase (which we provide) or singly using the wild type Cas9 nuclease, offering customers great flexibility. In addition, we also design truncated 17-, 18- and 19- mer sgRNAs for those customers who request it. We can also design sgRNAs pairs intended for use in the Cas9-FokI fusion approach.
Conclusions
Taken together, it’s clear that improvements in sgRNA design and strategy, combined with alternative versions of the Cas9 nuclease, make CRISPR-Cas9 a viable approach for many applications, including gene therapy, with much less concern about off-target mutagenesis than initially thought.
At GeneCopoeia, we offer a full suite of CRISPR-Cas9 genome editing tools, including both the wild-type and D10A nickase versions of Cas9. Our service includes sgRNA design for single and double nickase targets. We also carry sgRNA and Cas9 vectors in both lentiviral and non-viral formats. Finally, our services don’t stop at plasmid design and construction. We also generate stable cell lines and mouse lines carrying CRISPR-Cas9-mediated modifications, to meet all your genome editing needs.
References
Bogdanove & Voytas (2011). TAL Effectors: Customizable proteins for DNA targeting. Science 333, 1843.
Fu, et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature Biotechnology 31, 822.
Fu, et al. (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnology 32, 279.
Jinek, et al. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816.
Li, et al. (2013). Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 31, 681.
Mali, et al. (2013). Cas9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnology 31, 833.
Ran, et al. (2013a). Genome engineering using the CRISPR-Cas9 system. Nat. Protocols 8, 2281.
Ran, et al. (2013b). Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell 154, 1380.
Shalem, et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84.
Shen, et al. (2014). Efficient genome modification by CRISCRISCRISCRISPR-Cas9 nickase with minimal off target effects. Nature Methods 11, 399.
Tsai, et al. (2014). Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing.
Nature Biotechnology Apr 25. doi: 10.1038/nbt.2908. [Epub ahead of print].
Wang, et al. (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80.
Wu, et al. (2013). Correction of a Genetic Disease in Mouse via Use of CRISPR-Cas9. Cell Stem Cell 13,
659.
Xiao, et al. (2014). CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 30, 1180.
Yang, et al. (2013). One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas- mediated genome engineering. Cell 154, 1370.
| Copyright ©2014 GeneCopoeia, Inc. www.genecopoeia.com TNGE2-050814 |