Introduction
Biotin-protein ligase (EC 6.3.4.15) activates biotin to form biotinyl 5′ adenylate and transfers the biotin to biotin-accepting proteins. It also functions as a biotin operon repressor. The protein is encoded by the birA gene. Other names for this enzyme include: biotin ligase; biotin operon repressor protein; birA; biotin holoenzyme synthetase; biotin-[acetyl-CoA carboxylase] synthetase.
Source: E. coli
Storage buffer: 50 mM imidazole, pH 6.8, 50 mM NaCl, 5% glycerol, 5 mM mercaptoethanol
Storage conditions: The enzyme arrives on dry ice and should be immediately stored at -80°C. After the vial is thawed it should be stored at 4°C if it is to be re-used in the near future. For longer term storage a vial of thawed enzyme can be safely re-frozen by dropping into liquid nitrogen before storing at -80°C. Biomix A, B, and d-biotin can be stored at -20°C. There is no problem in thawing and re-freezing these mixtures.
Stability: Biotin ligase retains >90% of its activity for >3 months when stored at 4°C
Concentration: 1.0 mg/mL by A280
Purity: >98% by Coomassie staining
Activity: 5,000 Units/µg
Definition of Activity: 1 Unit is the amount of enzyme that will biotinylate 1 pmol of peptide substrate* in 30 minutes at 30°C using the reaction buffers provided and 38 µM peptide substrate*.
* The peptide substrate used in the enzyme assays was a 15-mer variant of sequence #85 identified by Schatz (1).
Contaminating proteases: <0.01% as chymotrypsin-like activity
Instructions for Use
Components provided
- Biomix-A (10X concentration: 0.5 M bicine buffer, pH 8.3)
- Biomix-B (10X concentration: 100 mM ATP, 100 mM MgOAc, 500 µM d-biotin)
- BirA enzyme
- Additional d-biotin
The final reaction mixture should contain: 1 part Biomix-A, 1 part Biomix-B and 8 parts substrate solution.
The amount of birA enzyme to add to the reaction mix may need to be varied to achieve biotinylation within a reasonable time-frame (see below). Typically, for every 10 nmol of substrate (at 40 µM), we recommend 2.5 µg of birA enzyme to complete the biotinylation in 30 – 40 min. at 30°C.
 |
 |
|
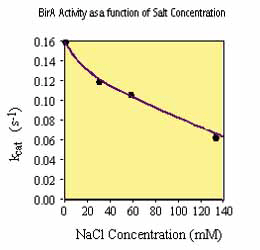
It should be noted that various reagents commonly present in biological buffers can inhibit the activity of birA enzyme including NaCl (100 mM), glycerol (5%) and ammonium sulfate (50 mM). Consequently, the concentration of these reagents in the substrate solution should be minimized. We recommend that, if possible, the substrate should be added to the reaction mix in 10 mM Tris-HCl, pH 8.
|
 |
|
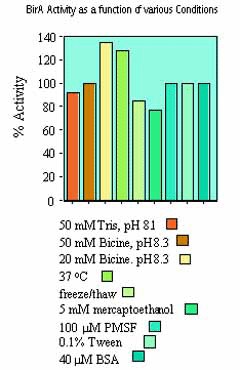
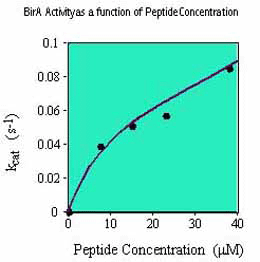
To ensure a rapid rate of biotinylation, it is recommended that the substrate be as concentrated as possible in the final reaction mix (up to 40 µM). The lower the substrate concentration in the reaction mix, the longer it will take to biotinylate. For example, whereas a substrate at 40 µM may be biotinylated in ~30 min., at 4 µM it will take ~5 hrs using the same amount of birA enzyme. To perform the biotinylation in 30 min. (i.e. 10 times faster), it is necessary to add 10 times more enzyme to the reaction mix.
|
 |
Biomix-A and -B have been optimized for the biotinylation of substrates at concentrations of no more than 40 µM. If it is desired to biotinylate substrate at concentrations of 40 – 80 µM, then it is necessary to supplement the reaction mix with additional biotin as follows: 1 part Biomix-A, 1 part Biomix-B, 7 parts substrate solution, 1 part supplemental biotin. For substrate concentrations above 80 µM, please contact Technical support at 301-762-0888 (866) 360 9531.
The reaction conditions described above have been optimized for a 15-mer peptide similar to sequence #85 identified by Schatz (1). The optimum reaction conditions for substrates in which the biotin peptide tag is attached to a protein was investigated and found that they are identical to the reaction conditions for the peptide substrate.
Bacterial Strains
Strain AVB 100
AVB100 is an E. coli K12 strain [MC1061 araD139 delta(ara-leu)7696 delta(lac)l74 galU galK hsdR2(rK-mK+) mcrB1 rpsL(Strr)] with a birA gene stably integrated into the chromosome.
Overexpression of the BirA protein is accomplished by induction with L-arabinose. The stably integrated birA gene does not require antibiotics to be maintained, and use of AVB100 with IPTG-inducible vectors such as Avidity’s pAC, pAN, pATN and pATC AviTag vectors allows independent control over the expressed gene of interest and the BirA levels. Use of this strain is limited to site of purchase. Please review the license included before using this product.
Strain AVB99
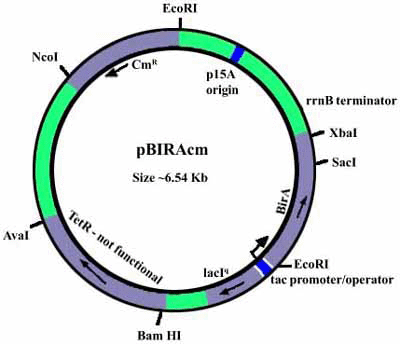
Strain AVB99 is an E.coli strain (XL1-Blue) containing a pACYC184 plasmid with an IPTG-inducible birA gene to overexpress biotin ligase (pBirAcm). The plasmid should be maintained with chloramphenicol at 10 µg/mL concentration (this is a low copy-number plasmid).
Strain AVB101
Strain AVB101 is an E. coli B strain (hsdR, lon11, su1A1), containing a pACYC184 plasmid with an IPTG-inducible birA gene to overexpress biotin ligase (pBirAcm). This strain is recommended for protein expression because of its robust growth and the absence of the OmpT and Lon proteases.
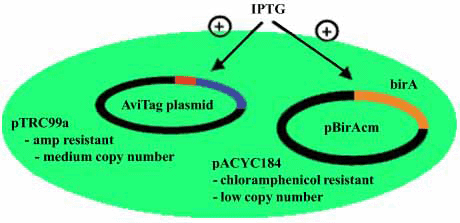
Expression of both biotin ligase and the AviTag protein are induced with IPTG (1 mM). Biotin should be added at the time of induction to a concentration of 50 µM. The pAN and pAC vectors require ampicillin for maintenance and the recommended concentration is 100 µg/mL.
Plasmid in Avidity’s AVB99 & AVB101
- The backbone is pACYC184.
- 1.3 Kb fragment contains lacIq inserted at EcoRV site (thus disrupting tetR gene).
- The BirA is expressed from the tac promoter operator.
- The rrnB terminator construct is between the laqIq region and destroys XbaI site of pACYC184.
|
 |
Publications
View selected publications citing GeneCopoeia’s Biotin Protein Ligase products from the recent literature
- Brouwer, P.J.M., et al. (2021). Two-component spike nanoparticle vaccine protects macaques from SARS-CoV-2 infection. Cell doi: 10.1016/j.cell.2021.01.035 [Biotin protein ligase]
- Guo, Y., et al. (2021). A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes. Nature Communications doi: 10.1038/s41467-021-22926-2 [Biotin protein ligase]
- Dai, L., et al. (2021). Protective Zika vaccines engineered to eliminate enhancement of dengue infection via immunodominance switch. Nature Immunology doi: 10.1038/s41590-021-00966-6 [Biotin protein ligase]
- Xie, Y., et al. (2021). Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1. Cell Reports doi: 10.1016/j.celrep.2021.109600 [Biotin protein ligase]
- Gao, X-Y., et al. (2021). Development of Fc-specific multi-biotinylated antibodies via photoreactive tandem AviTag repeats for the ultrasensitive determination of ochratoxin. Food Control doi: 10.1016/j.foodcont.2021.108525 [Biotin protein ligase]