Introduction
Introduction
NGS technology is widely used in drug discovery and translational medicine, such as determining individual genome sequences and confirming mutations associated with genetic diseases and somatic mutations in tumour cells. Parallel assay reference standards are commonly used to assess the sensitivity and reproducibility of each instrument and kit for the detection of mutations at each specific locus.
However, parallel reference standards cannot be used to assess and correct the number of molecules containing mutations including substitutions, deletions and insertions in the samples to be examined, nor can they be used to exclude errors in a large number of samples to be examined due to a certain experimental step (e.g., library construction, efficiency of barcode ligation, or loss of part of the samples in a certain sample system due to operational errors) or instrumental heterogeneity (e.g., abnormalities of a certain well of a 96-well PCR instrument). (e.g., an abnormality in one well of the 96-well PCR instrument).
GeneCopoeia provides
OncoSpot™ NGS Spike-in Reference Standards for monitoring the entire process of detection. The CRISPR method is employed to modify the genome of tumour cells, introducing single nucleotide mutations or in-frame indel mutations. Concurrently, spike-in markers are set up using our patented technology, namely the ‘DNA reference standard and use thereof’.
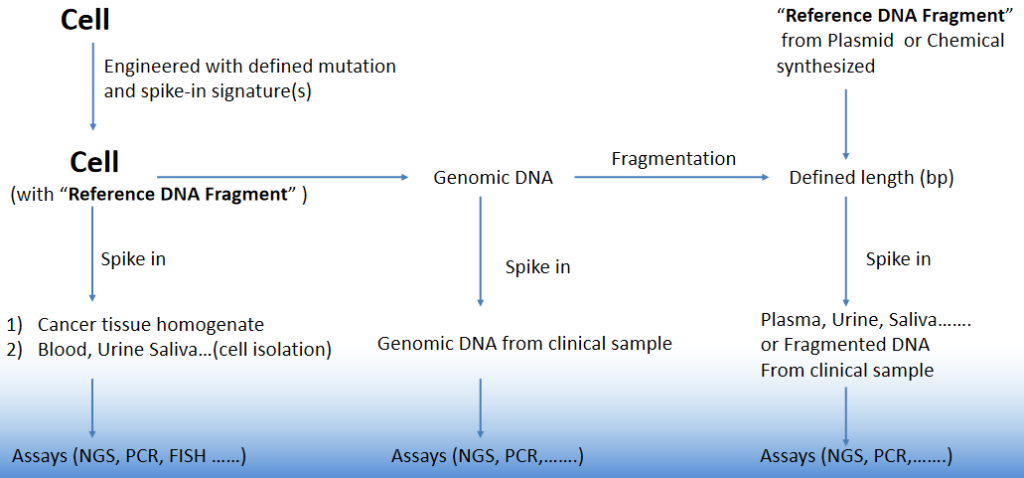
Fig. 1 Cell/gDNA/cfDNA spike-in reference standards for gene mutation detection
Advantage
- Adding to the actual sample as an internal reference, the Spike-in Reference Standards are able to participate in all the processes of DNA library sequencing, and take quality control at each processes.
- The sequencing results of different samples can be normalized based on the results of the Spike-in Reference Standards, allowing for comparison between samples;
- Highly similar to the form of the real sample, but can be easily distinguished;
- Flexible customization, which can meet the needs of a variety of quality control indicators.
Application
Application
- Evaluate the efficiency of each step of sample detection, including samples collection, preservation, and nucleic acid extraction methods. And evaluate the effectiveness of the combination of quality controls and reference standards.
- Assessing the quality of sample collection.
- Assessing the sensitivity and specificity of enzymes and buffers in reaction systems.
- Evaluate the temperature control of the wells in which the instrument samples (negative tubes) are placed for testing and provide a reference for instrument service engineers.
- As a basis for calculating the correction factor for the number of copies of DNA or RNA in the sample (copies/mL).
Example
cfDNA Spike-in Reference Standard (Lung Cancer Mutation Sites)
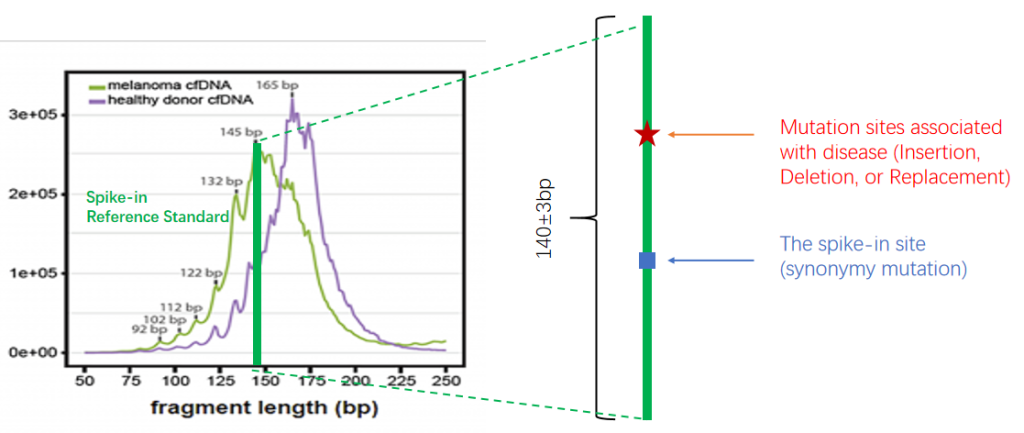
Fig.2 Difference in length of cfDNA between cancer patient and normal human.
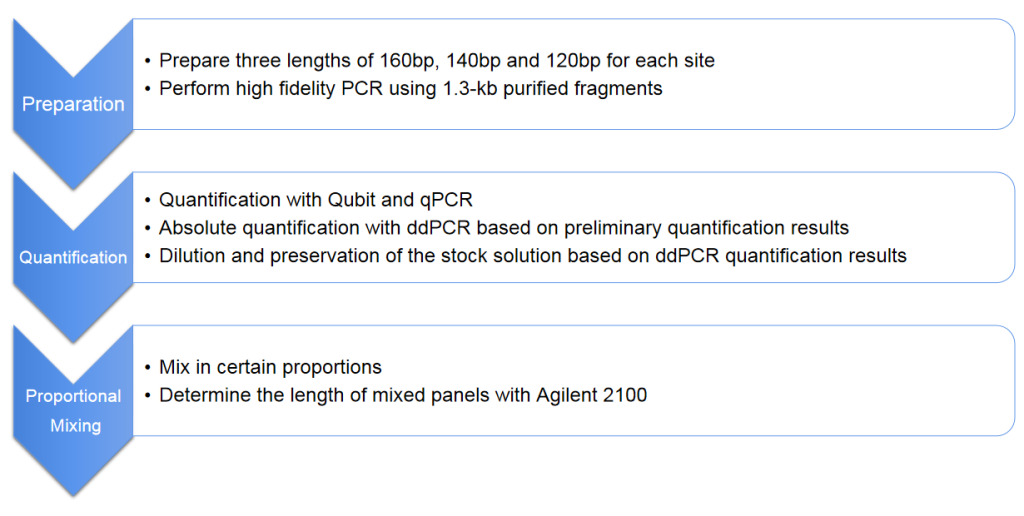
Fig.3 Procedure for the preparation of Spike-in Reference Standard for lung cancer mutant cfDNA.
Order
| Product |
Gene |
Variant |
Price |
| KRAS G13C Spike-in Reference Standard |
KRAS |
G13C |
Inquire |
| BRAF V600E Spike-in Reference Standard |
BRAF |
V600E |
Inquire |
| EGFR Δ746-750 Spike-in Reference Standard |
EGFR |
Δ746-750 |
Inquire |
| CKIT D816V Spike-in Reference Standard |
CKIT |
D816V |
Inquire |
| PIK3CA E545K Spike-in Reference Standard |
PIK3CA |
E545K |
Inquire |
| EGFR G719S Spike-in Reference Standard |
EGFR |
G719S |
Inquire |
| NRAS Q61K Spike-in Reference Standard |
NRAS |
Q61K |
Inquire |
| EGFR L858R Spike-in Reference Standard |
EGFR |
L858R |
Inquire |
| PIK3CA H1047R Spike-in Reference Standard |
PIK3CA |
H1047R |
Inquire |
| EGFR T790M Spike-in Reference Standard |
EGFR |
T790M |
Inquire |
We can provide customised Spike-in Reference Standards (e.g. in the form of simulated viruses, cells, cfDNA, etc.), or we can provide the technology to Spike-in Reference Standards related projects. Please
contact us if you are interested!
FAQs
1.What is spike-in reference standard?
A:Spike-in Reference Standards refer to positive standards that can be incorporated into the sample to be tested or into the sample collection device (tube). During sample processing, the standard needs to be added to the sample collection device (tube) and processed together with the sample to be tested, so as to evaluate the stability of the testing process and the accuracy of the test results of other samples based on the test results of the standard.
2. What is the difference between Spike-in Reference Standards and reference standards for parallel experiments?
A:At present, the reference standards for parallel experiments used in NGS can only be used to analyze and correct the systematic error of the experiment. Including: 1) to compare and evaluate the relevant instruments and equipment, kits, operators, laboratory conditions and quality of a certain testing project; 2) for quality analysis and optimisation of the individual experimental (testing) steps of an assay.
However the Spike-in Reference Standards are not only act as reference standards for parallel experiments, but also 1) participate in all processes of DNA library sequencing and quality control at every step of the process; 2) the sequencing results of different samples can be normalized according to the results of the spike-in reference standards, so as to make comparisons between samples.
3. How to prepare Spike-in Reference Standard?
A:GeneCopoeia modifies the genome of tumour cells by CRISPR, or by PCR amplification, introducing single nucleotide mutations or in-frame indel mutations, and at the same time sets up spike-in markers with our own patented technology ‘DNA reference standard and use thereof’. The Spike-in Reference Standards are capable of monitoring the whole process of the detection.




