GeneCopoeia's GeneHero™ CRISPR-Cas9 products and services provide a complete, powerful solution to your genome editing needs. Products and services include:
- CRISPR Plasmids
Transfect cells with our CRISPR plasmids with Cas9 and sgRNA for human, mouse, and rat. Search our database of more than 45,000 human, mouse, and rat genes for genome editing using CRISPR.
- CRISPR Lentivirus
Genome integration of CRISPR elements using lentivirus. Cas9 and/or sgRNA packed in purified lentiviral particles at 108TU/ml, ready to infect all cell types.
- CRISPR AAV
Episomal expression of CRISPR components with adeno-associated viral particles carrying Cas9 and/or sgRNA, excellent for tissue and animal transduction.
Premade Cas9-expressing stable cell lines are great for sgRNA library screening and other high-throughput CRISPR-Cas9 applications.
Advantages
- RNA-guided genomic DNA recognition regardless of the methylation status
- Similar or greater gene-editing efficiency compared to ZFNs and TALENs
- Capable of editing multiple genes simultaneously (multiplexing)
- Simple and fast design process. No need to reengineer the nuclease for each new target
sgRNA expression plasmids are available for targeting virtually any gene in any experimental system. sgRNA plasmids express either sgRNA only, or sgRNA + Cas9 nuclease in an all-in-one format. Lentiviral clones express sgRNA alone. Cas9-expressing lentiviral clones are available separately.
The CRISPR plasmid options listed below are intended for using 20 ntsgRNAs with the wild-type Cas9 nuclease from Streptococcus pyogenes(SpCas9). GeneCopoeia also provides alternate CRISPR options, such as Cas9 D10A nickase and other high-fidelity versions of Cas9, SaCas9, and shorter sgRNAs. To purchase reagents for these other options, please contact us for a custom quote.
Custom sgRNA Expression Plasmids
Cas9 Nuclease Expression Plasmids
Clones expressing human codon-optimized S. pyogenes Cas9 nuclease or D10A nickase are available. These clones do not express sgRNAs.
Click here to search our database of sgRNA plasmids for knocking out more than 45,000 human, mouse, and rat genes.
Custom sgRNA Lentiviral Particles
sgRNA for human, mouse, and rat packaged in high-titer lentivirus.
| Vector | Promoter | sgRNA and Cas9 | Selection Marker/ Reporter Gene | Vector Type | Vector Map |
|---|---|---|---|---|---|
| pCRISPR-LvSG03 | U6 | sgRNA only | Puromycin / mCherry | Lentiviral |
Pre-made sgRNA Control Particles
Pre-made Cas9 Nuclease Lentiviral Particles
sgRNA and Cas9 AAV Particles
| Vector name | CRISPR | Description | Reporter gene |
|---|---|---|---|
| pCRISPR-AD01 | SaCas9 & sgRNA | All-in-one AAV vector with SaCas9 and sgRNA | N/A |
| pCRISPR-AV20 | sgRNA only | AAV vector expressing sgRNA driven by U6 promoter | N/A |
The clustered, regularly interspaced, short palindromic repeats (CRISPR) system is bacterial immunity mechanism for defense against invading viruses and transposons. This system has been adapted for highly efficient genome editing in many organisms. Compared with earlier genome editing technologies such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), CRISPR-Cas–mediated gene targeting has similar or greater efficiency. Genome editing has been used for numerous applications, as shown in Table 1.
Table 1. Applications for CRISPR-mediated genome editing.
| Application | Description |
|---|---|
| Gene knockout | Permanently modify DNA to eliminate gene function. |
| Gene mutagenesis | Introduce point mutations to an endogenous gene |
| Gene tagging | Add a fusion tag (e.g. luciferase, GFP) to track an endogenous promoter activity or an endogenous protein expression and location |
| Safe harbor knock-in | Knock-in an exogenous ORF or other genetic element to safe harbor sites of human or mouse genome |
| Gene activation | Activate an endogenous gene expression |
| Gene repression | Repress an endogenous gene expression |
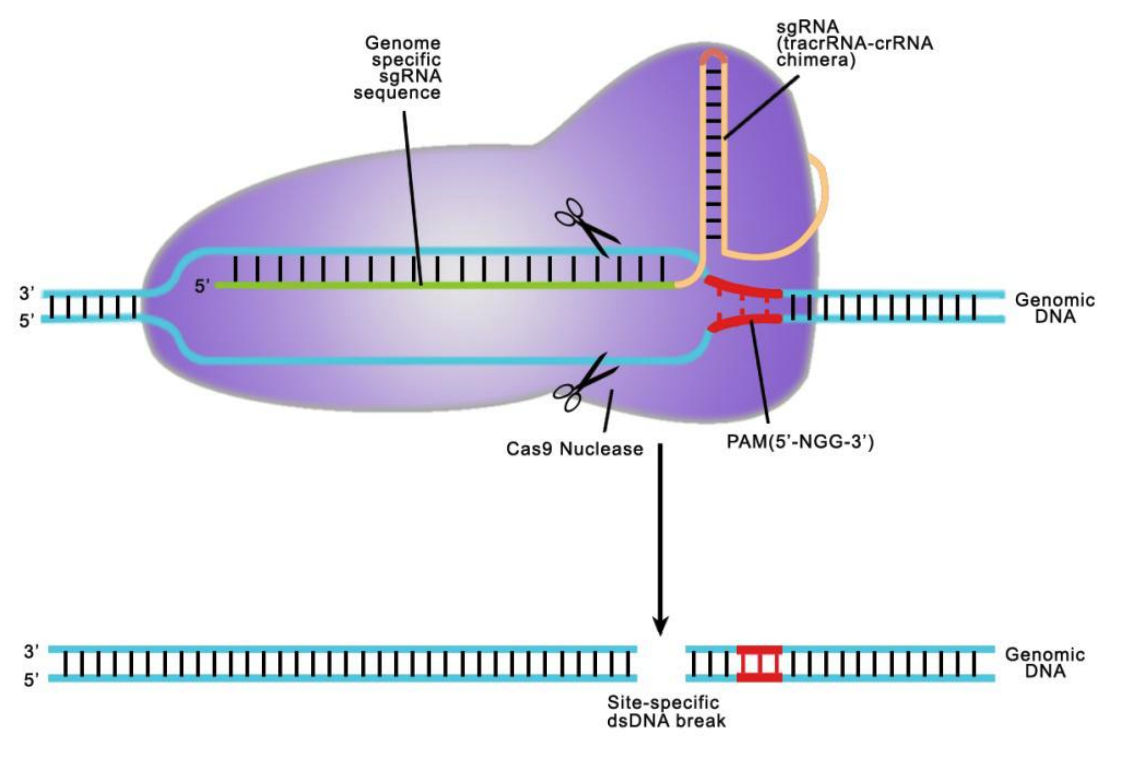
In the type II CRISPR systems, the complex of a CRISPR RNA (crRNA) annealed to a trans-activating crRNA (tracrRNA) guides the Cas9 endonuclease to a specific genomic sequence, thereby generating double-strand breaks (DSBs) in target DNA. This system has been simplified by fusing crRNA and tracrRNA sequences to produce a synthetic, chimeric single-guided RNA (sgRNA). The sgRNA contains within it a 20 nucleotide DNA recognition sequence (Figure 1).
Figure 1. Mechanism of CRISPR-Cas9-sgRNA target recognition and cleavage.
When the Cas9-sgRNA complex encounters this target sequence in the genome followed by a 3 nucleotide NGG PAM (protospacer adjacent sequence) site, the complex binds to the DNA strand complementary to the target site. Next, the Cas9 nuclease creates a site-specific double-strand break (DSB) 3-4 nucleotides 5' to the PAM. DSBs are repaired by either non-homologous end joining (NHEJ), which is error-prone, and can lead to frameshift mutations, or by homologous recombination (HR) in the presence of a repair template (Figure 2).
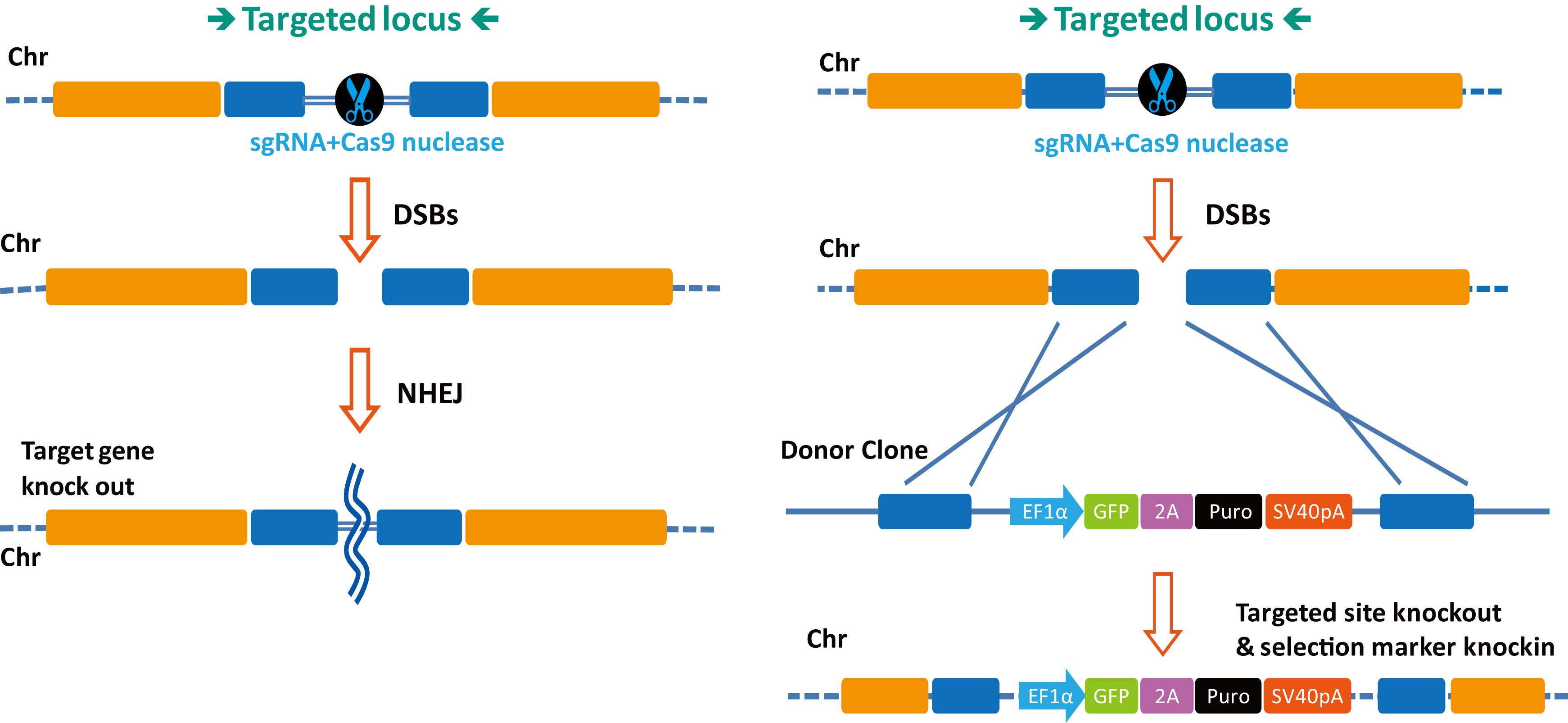 |
Figure 2. CRISPR-Cas9-based gene engineering. Left. DSBs created by sgRNA-guided Cas9-mediated cleavage are repaired by NHEJ. Right. DSBs created by sgRNA-guided Cas9 nuclease is repaired homologous recombination between sequences flanking the DSB site, thereby causing "knock-in" of sequences on a donor DNA.
While the CRISPR system provides a highly efficient means for carrying out genome editing applications, it is prone to causing off-target indel mutations. Off-targeting is caused by the ability of the Cas9- sgRNA complex to bind to chromosomal DNA targets with one or more mismatches, or non-Watson-Crick complementary. The propensity of CRISPR for off-target modification is a significant concern for some researchers who want to avoid results that are potentially confounded by off-target modification, as well as for those who might be interested in developing CRISPR for gene therapy applications.
Several strategies have been employed to mitigate CRISPR's propensity for off-target genome modification. One such strategy is to use double nickases to create DSBs. The Cas9 D10A mutant is able to cleave only one DNA strand, thereby creating a "nick". When two sgRNAs that bind on opposite strands flanking the target are introduced, two Cas9 D10A nickase molecules together create a staggered-cut DSB, which is then repaired by either NHEJ or HR (Figure 3). The double nickase strategy has been shown to greatly reduce the frequency of off-target modification. However, double nickases are limited in utility by design constraints; the sgRNAs must be on opposite strands, in opposite orientation to one another, and display optimal activity when spaced from 3-20 nucleotides apart. In addition, the cleavage activity of double nickases tends to be lower than that of standard Cas9-sgRNA. Further, nickases can still cause some degree of off-target indel formation.
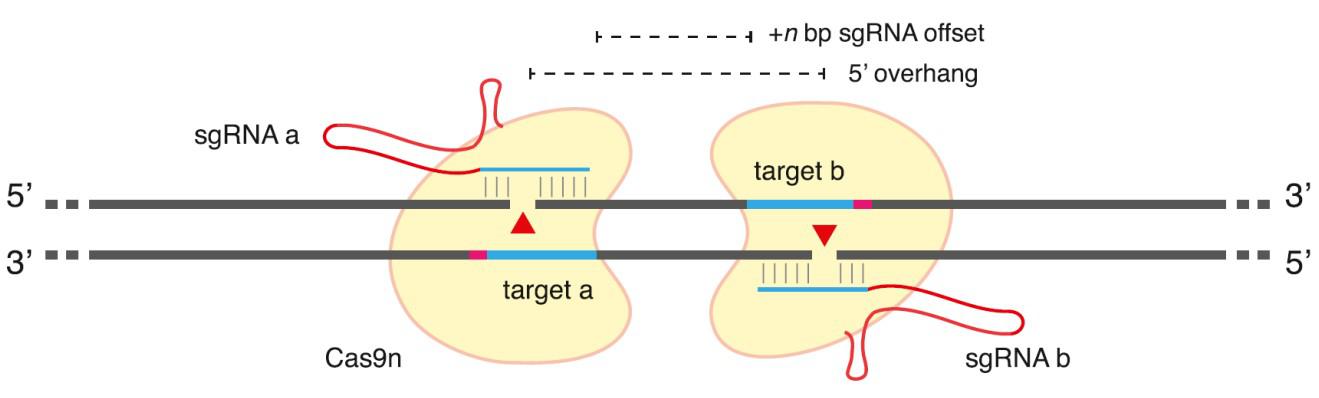 Figure 3. General scheme of Cas9 double-nickase strategy. From Ran, et al. (2013). Two additional strategies, the use of truncated (17-18 nucleotide) sgRNAs, as well as a Cas9-FokI fusion, also dramatically reduce CRISPR-mediated off-target genome modification. However, these methods suffer from even further reductions in on-target activity and/or more severe design constraints compared with the double nickase approach.
Figure 3. General scheme of Cas9 double-nickase strategy. From Ran, et al. (2013). Two additional strategies, the use of truncated (17-18 nucleotide) sgRNAs, as well as a Cas9-FokI fusion, also dramatically reduce CRISPR-mediated off-target genome modification. However, these methods suffer from even further reductions in on-target activity and/or more severe design constraints compared with the double nickase approach.
Recently, two groups demonstrated that engineering Cas9 variants carrying 3-4 amino acid changes virtually eliminates CRISPR off-target genome modification. These variants still retain high on-target activity, without the design constraints of previous approaches, providing a promising alternative for high-fidelity CRISPR-mediated genome editing.
References
- Horvath P, Barrangou R (January 2010). "CRISPR/Cas, the immune system of bacteria and archaea". Science 327 (5962): 167–70.
- Marraffini LA, Sontheimer EJ (February 2010). "CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea". Nat Rev Genet 11 (3): 181–190.
- Hale CR, Zhao P, Olson S, et al. (November 2009). "RNA-Guided RNA Cleavage by a CRISPR RNA-Cas Protein Complex". Cell 139 (5): 945–56.
- van der Oost J, Brouns SJ (November 2009). "RNAi: prokaryotes get in on the act". Cell 139 (5): 863–5. doi:10.1016/j.cell.2009.11.018.
- Hale CR, Zhao P, Olson S, et al. (November 2009). "RNA-Guided RNA Cleavage by a CRISPR RNA-Cas Protein Complex". Cell139 (5): 945–56.
- Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J.A., and Charpentie E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptiv bacterial immunity. Science 337, 816–821.
- Jiang, W., Bikard, D., Cox, D., Zhang, F., and Marrafï¬ni, L.A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat.Biotechnol. 31, 233–239.
- Hsu, P.D., Scott, D.A.,Weinstein, J.A., Ran, F.A., Konermann, S., Agarwala, V.,Li, Y., Fine, E.J., Wu, X., Shalem, O., et al. (2013). DNA targeting speciï¬city of RNA-guided Cas9 nucleases. Nat. Biotechnol. Published online July 21, 2013.
- Ran, et al. (2013). Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell 154, 1380.
- Knockout By TALEN Or CRISPR vs. Knockdown By shRNA or siRNA
- CRISPR-Cas9 Specificity: Taming Off-target Mutagenesis
- Genome Editing: Which Should I Choose, TALEN Or CRISPR?
- Genome Editing In Mammalian Cells: What Do I Do Next?
- Genome Editing: HDR Donors For Gene Knockout, Mutagenesis, Tagging, and Safe Harbor Knock-in
- Genome Editing: Applications For GeneCopoeia CRISPR sgRNA Libraries
- IndelCheck™: A Powerful CRISPR/TALEN Validation & SCreening Tool
- Genome Editing: Cas9 Stable Cell Lines for CRISPR sgRNA Validation, Library Screening, and More
- CRISPR-Cas9 Protein for Highly Efficient Genome Editing With Minimal Off-Target Activity
- Why You Should Know Your Gene's Accession Number
Watch recorded webinar / Download slides Title: Strategies For Effective CRISPR-Mediated Gene Modification Presented Monday, December 3, 2018
CRISPR genome editing-the ability to make specific changes at targeted genomic sites-has transformed research in biology and medicine, due to its precision, ease of use, and relatively low cost. CRISPR's many applications include gene knock out, gene tagging, correction of genetic defects, and gene activation. In this webinar, we discuss how to use GeneCopoeia’s CRISPR technology for efficient genome editing in mammalian cell lines.
Watch recorded webinar Download slides Title: Harnessing CRISPR for Activation of Gene Expression Presented Wednesday, November 14, 2018
CRISPR-Cas systems have been successfully re-purposed from bacterial adaptive immune mechanisms for the modification of genes. Most CRISPR-mediated gene modifications include permanently changing the genetic code for such applications as gene knockout, mutagenesis, and fusion tagging. However, CRISPR-Cas has also been adapted for stimulating gene transcription, without modifying coding sequences. In this webinar, we discuss how GeneCopoeia CRISPR products and services can help you get more from your transcriptional activation applications.
Watch recorded webinar / Download slides Title: Applications For CRISPR-Cas9 Stable Cell Lines Presented Wednesday, March 22, 2017
The CRISPR-Cas9 system has become greatly popular for genome editing in recent years, due to its ease-of-design, efficiency, specificity, and relatively low cost. In mammalian cell culture systems, most genome editing is achieved using transient transfection or lentiviral transduction, which works well for routine, low-throughput applications. However, for other applications, it would be beneficial to have a system in which one component, namely the CRISPR-Cas9 nuclease, was stably integrated into the genome. In this webinar, we introduce GeneCopoeia’s suite of Cas9 stable cell lines, and discuss the great utility that these cell lines provide for genome editing applications.
Watch recorded webinar / Download slides Title: Safe Harbor Transgenesis in Human & Mouse Genome Editing Presented Wednesday, April 19, 2017
Insertion of transgenes in mammalian chromosomes is an important approach for biomedical research and targeted gene therapy. Traditional lentiviral-mediated transgenesis is effective and straightforward, but its random integration can often be unstable and harm cells. "Safe Harbor" sites in human and mouse chromosomes have been employed recently as an alternative to random, viral-mediated integration because they support consistent, stable expression, and are not known to hamper cell fitness or growth. In this webinar, we will discuss the merits of Safe harbor transgenesis approaches, and how GeneCopoeia's CRISPR tools for Safe Harbor knock-in can greatly benefit your research.
Watch recorded webinar / Download slides Title: GeneCopoeia CRISPR sgRNA Libraries For Functional Genomics Presented Wednesday, April 29, 2015
Biomedical researchers are enjoying a Renaissance in functional genomics, which aims to use a wealth of DNA sequence information—most notably, the complete sequence of the human genome—to determine the natural roles of the genes encoded by the genome. As a result, biochemical networks and pathways will be better understood, with the hope of leading to improved disease treatments. Researchers are turning increasingly to CRISPR (clustered, regularly interspaced, short palindromic repeats) for functional genomics studies. Several groups recently adapted CRISPR for high-throughput knockout applications, by developing large-scale CRISPR sgRNA libraries. GeneCopoeia recently launched a number of smaller, pathway- and gene group-focused CRISPR sgRNA libraries, which offer several key advantages over the whole-genome libraries. In this 40 minute webinar, we discuss the merits and applications for CRISPR sgRNA libraries, how to use CRISPR sgRNA libraries, the advantages of using small, pathway- and gene group-focused libraries, and how GeneCopoeia can help you with your high-throughput CRISPR knockout studies.
GeneCopoeia recommends our donor plasmid design and construction service. We will construct a donor plasmid that contains a defined modification, flanked by a selectable marker such as puromycin resistance, and homologous arms from your target region. The donor may or may not also include a fluorescent reporter such as GFP. The markers can be flanked by loxP sites, to permit Cre-mediated removal, if desired. Use of a GeneCopoeia-designed donor plasmid allows you to select for edited clones and reduces the number of clones required for screening. You can also purchase our donor cloning vectors for do-it-yourself donor clone construction.
GeneCopoeia recommends our donor plasmid design and construction service. We will construct a donor plasmid that contains a defined modification, flanked by a selectable marker such as puromycin resistance, and homologous arms from your target region. The donor may or may not also include a fluorescent reporter such as GFP. The markers can be flanked by loxP sites, to permit Cre-mediated removal, if desired. Use of a GeneCopoeia-designed donor plasmid allows you to select for edited clones and reduces the number of clones required for screening. You can also purchase our donor cloning vectors for do-it-yourself donor clone construction.
1. Standard transfection. GeneCopoeia recommends our EndoFectin™ transfection reagents. Transfection can be used for either DNA, RNA, or Cas9 nuclease protein.
2. Electroporation, which can be used for either DNA, RNA, or Cas9 nuclease protein.
3. Lentiviral transduction (cannot be used with HDR donors).
4. Micro-injection of either DNA, mRNA, .or Cas9 nuclease protein. The plasmids can be used as templates for in vitro transcription of RNA, or synthetic RNA can be used.
1. If you are making an insertion or deletion, the easiest way to screen your cells is by PCR using primers flanking the modified site, provided that the insertion or deletion is large enough to detect by standard agarose gel electrophoresis.
2. For very small insertions or deletions, you can screen your clones using GeneCopoeia's IndelCheck™ T7 endonuclease I assay, which is a method that detects mutations by cleaving double stranded DNA containing a mismatch. You can also screen using Sanger sequencing of PCR products. Ultimately, Sanger sequencing needs to be done to verify the presence of the modification.
3. If you are introducing a point mutation, then you can use either real-time PCR or Sanger sequencing to detect the modification.
4. If the modification you are introducing creates or destroys a restriction enzyme site, then enzyme cleavage of PCR products can be used to distinguish between modified and unmodified alleles.
5. Finally, either Sanger sequencing of PCR products or Next Generation sequencing of whole genomes can be used to screen for modifications. Regardless of which screening method you choose, it is also important that you are able to determine whether only a portion or all of the alleles have been modified.
In order to reduce the amount of time and effort required to identify edited clones, GeneCopoeia recommends our donor plasmid design and construction service. We will construct a donor plasmid that contains a defined modification, flanked by a selectable marker such as puromycin resistance, and homologous arms from your target region. The donor may or may not also include a fluorescent reporter such as GFP. The markers can be flanked by loxP sites, to permit Cre-mediated removal, if desired. Use of a GeneCopoeia-designed donor plasmid allows you to select for edited clones and reduces the number of clones required for screening. You can also purchase our donor cloning vectors for do-it-yourself donor clone construction.