| Ed Davis, Ph.D. |
Introduction
Fusion tagging-the practice of adding amino acids in-frame to one end of a native protein-is a well- established strategy for many applications, including protein purification, immunoprecipitation, Western blotting, and in vivo imaging. GeneCopoeia provides tens of thousands of human and mouse open reading frame (ORF) clones for ready expression in vitro and in vivo. These clones come either without tags or with a large number of fusion tag options for customers to choose from. However, designing a tagging strategy requires consideration of many factors, depending on the particular application and the goals of the experiment. In this Technical Note, we discuss the many tag choices available for GeneCopoeia ORF clones, and what you need to consider in order to make the best tag choice.
GeneCopoeia OmicsLink™ Expression-ready ORF clones for studying protein function
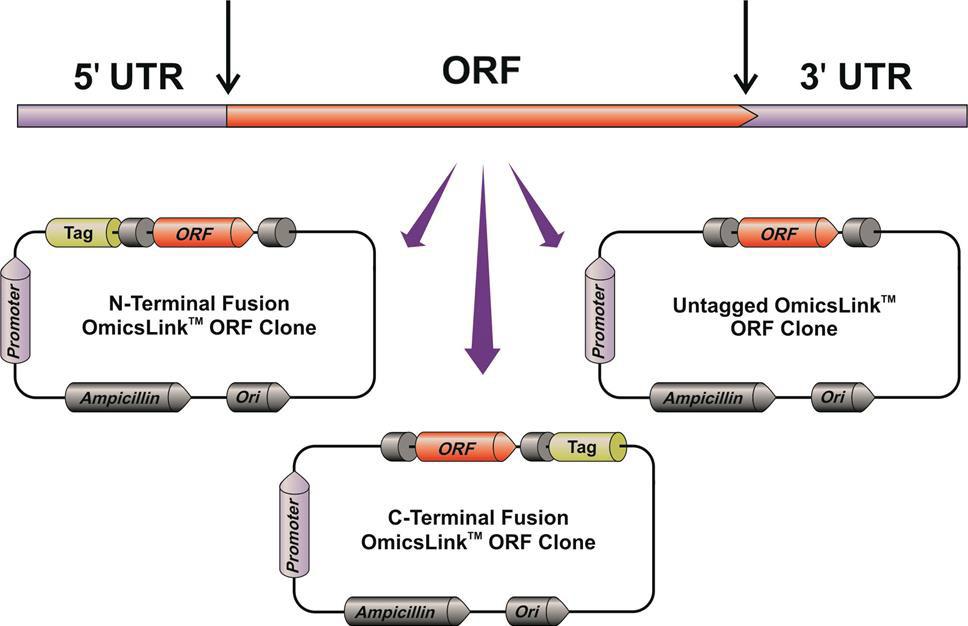
GeneCopoeia’s OmicsLink™ Expression-ready ORF clones carry human and mouse ORF sequences. ORFs are transferred to vectors from cDNAs, beginning with the initiator ATG and ending at the stop codon (the stop codon is removed when the ORF is connected to a C-terminal tag). The end result is that the natural 5’ and 3’ untranslated regions (UTRs) have been removed (Figure 1).
Figure 1. GeneCopoeia strategy for transferring ORF sequences from full-length cDNAs to ready-to-use expression vectors lacking the natural 5’ and 3’ UTRs.
In these vectors, ORFs acquire the following features:
- Expression is placed under control of various promoters, such as CMV, EF1, etc.
- Strong termination signals are added at the 3’ end, such as the SV40 polyA.
- ORFs come either untagged, for expression of the protein with its native structure, or with N- or C-terminal fusion tags.
Regardless of vector type, promoter, or tag, all GeneCopoeia OmicsLink™ Expression-ready ORFs are sequence-verified, and guaranteed to be free of PCR or cloning errors, frameshifts, premature stop codons, and single nucleotide polymorphisms (SNPs) not known to occur in nature.
Consideration 1: What’s my application?
While GeneCopoeia ORF clones come with a choice of more than 150 tags and tag combinations (Table 1), they can also be provided in untagged format. A chief advantage of untagged ORFs is that they provide the opportunity to study the protein with its native structure. However, if you require a tag, then the choice of tag will depend heavily on the application. Here are some examples:
Protein purification
One of the most common uses for tagging is protein purification, in which the tagged protein is usually expressed in E. coli. Bacterial lysates are run through an affinity column, which binds to the tagged protein. The tagged protein is eluted from the column in a highly purified form. Typically, the small 6XHis tag (six histidine residues) is used for purification of proteins from cells. The 6XHis tag is widely used for purification from E. coli, but it is not recommended for use in purification of non-secreted proteins from mammalian cells, due to a naturally high background of histidine. Instead, many researchers use another small tag, FLAG® (D-Y-K-D-D-D-D-K) for this purpose. If solubility is an issue, many researchers use the larger tags glutathione S-transferase (GST) or maltose binding protein (MBP).
Detection on a Western blot
If you need to use a Western blot to detect expression of your protein from cell lysates, then choose a small tag for which there is a good antibody. The FLAG® tag is a commonly-used tag for Western detection, due to its small size, and a prevalence of many good commercial antibodies for it. Other appropriate choices for fusion tags for Western blotting include 6XHis, C-Myc, HA, and HaloTag®.
Immunoprecipitation/pull-down
In some experiments, you might want to isolate your protein under native (non-denaturing) conditions. Or your protein might be part of a complex, and you want to isolate some or all of the complex’s components. Then choose a pull-down tag, which can be used for either Immunoprecipitation or chemical affinity. For Immunoprecipitation, cell lysates are mixed with an antibody attached to a solid support, like agarose or magnetic beads. The lysate is then centrifuged or placed in a magnetic field to separate the heavy complex-which contains the solid support, antibody, and protein-from the rest of the lysate. The FLAG® tag, for the same reasons as described above for Western blotting, is typically used for Immunoprecipitation. Other commonly-used tags for Immunoprecipitation are HA and cMyc.Alternatively, researchers use chemical affinity for pull-down. One example of a chemical affinity tag provided by GeneCopoeia is the AviTag™. AviTag™ is a small tag that binds with high affinity to biotin. Biotin itself can also be coupled to agarose or magnetic beads, and so, as with Immunoprecipitation, complexes consisting of the bead, biotin, and protein of interest can be separated from the lysates.
Live cell imaging
Fluorescent proteins (FPs) are most often used for live cell imaging. The most common FPs are green fluorescent protein (GFP) and its derivatives (CFP, YFP), and some red variants, such as dTomato and mCherry. FP tags enable viewing of living cells under a fluorescent microscope, or separation of live cells via fluorescence activated cell sorting (FACS) in real time and without any introduction of substrates.
HaloTag®, a small tag that binds many different ligands, is an alternative to FPs. Using one tag, researchers can add different ligands to cell media, each of which fluoresce at different wavelengths.
Many tags can be used for multiple applications. Some are more suitable for some applications than others. Table 1 displays many of the tags available for GeneCopoeia ORF clones and their applications.
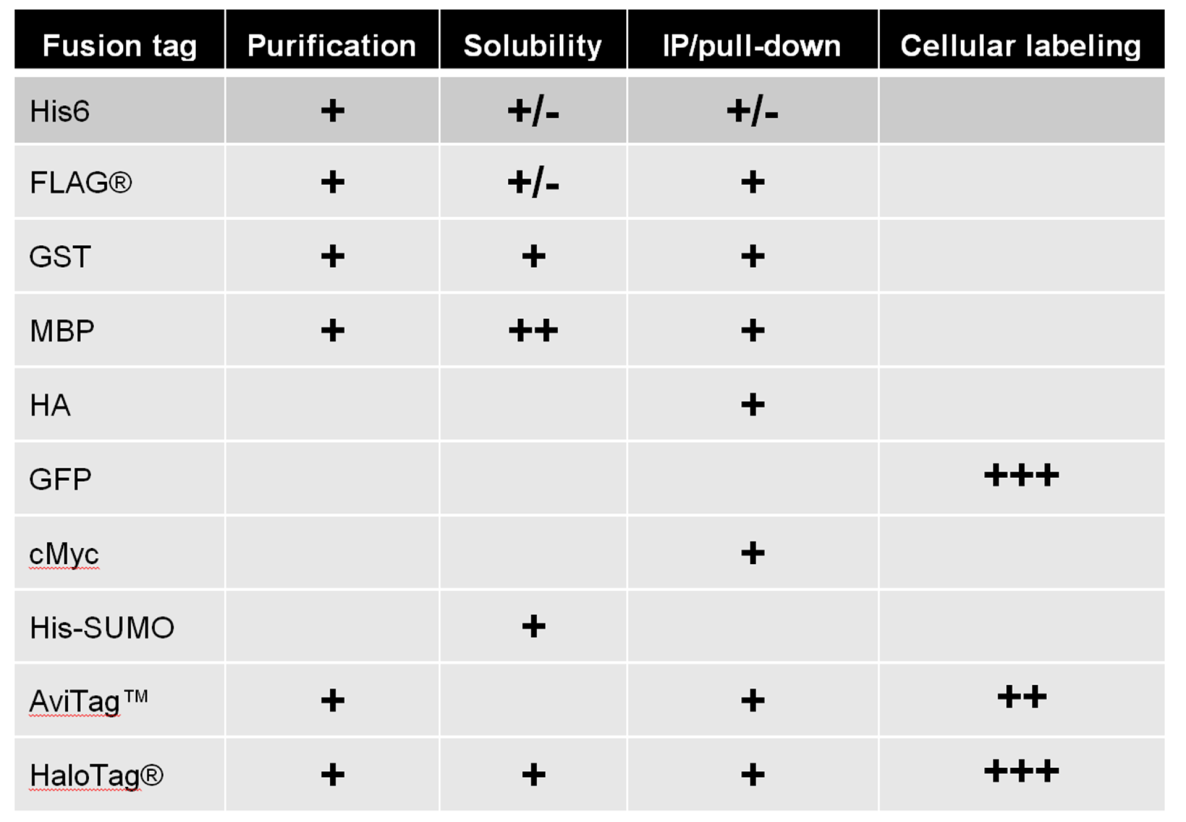
Table 1. Examples of tags available in GeneCopoeia OmicsLink™ Expression-ready ORF clone vectors.
Consideration 2: Impact of a fusion tag
Before making the decision to use a fusion tag, it is very important to understand that any kind of tag, in any position of the amino acid sequence, has the potential to have a profound impact on the expression or function of the protein you are interested in (Bucher, et al., 2002). One major concern is that addition of the tag could interfere with proper protein folding, rendering your protein of interest inactive or insoluble. A second major concern is that addition of a tag could disrupt a subcellular localization signal. In that case, the protein could be expressed and folded correctly, but it is in the wrong place in the cell.
In some cases, the interference of protein function by a tag can be alleviated by tag removal. Some tags in the GeneCopoeia vector arsenal come with the recognition site for Tobacco Etch virus (TEV) protease, which can be used in trans to remove an associated fusion tag (Kapust & Waugh, 2000). A similar protease cleavage site associated with some tags is Enterokinase (Ek; Terpe, 2003). GeneCopoeia’s CoolCutter™ is a small ubiquitin-like modifier (SUMO) protease that recognizes the tertiary structure of SUMO rather than a specific amino acid sequence. Using CoolCutter™ allows the SUMO tag to be removed, leaving only untagged protein with no remnants of a protease cleavage site.
A potentially negative impact posed by fusion tagging can be alleviated by separating the tag from the protein of interest in one of two ways. One of these uses an internal ribosome entry site (IRES), which is an element that, when inserted between two ORFs on the same transcript, allows independent translation of each protein (Kozak, 2005). However, use of an IRES for tagging comes with two caveats: 1) Such elements eliminate the goal of actual fusion tagging, instead creating two independent protein molecules: and 2) IRES elements do not always function as intended, so in some cases the protein either upstream or downstream of the IRES is translated at reduced levels (de Felipe, et al., 2006).
One can also separate two proteins from one another using 2A peptides, (Ryan, et al., 1991), which are inserted in-frame between the protein of interest and the tag. Thus, 2A peptides represent true fusion tagging. Once the bicistronic message is translated, the 2A peptide cleaves itself, separating the two proteins from one another. However, as with IRESs, the protein is no longer tagged, and the tag cannot be used for any of the aforementioned applications.
Consideration 3: N- or C-terminal?
The decision whether to choose the N- or C-terminus for tag placement is based on several factors. First, you should not choose an end that is buried in the core of the protein, because then it is not likely to be accessible to antibodies or other ligands in non-denaturing (i.e. native) conditions. Second, you should also avoid placing a tag near a functional domain, to minimize disrupting the protein’s natural activity.
Further, many proteins have N-terminal localization signals. Placing a tag at the N-terminus of a protein could interfere with the function of the signal sequence and lead to mislocalization. Alternatively, the localization sequence could be removed post-translationally, in which case the tag will be lost. Indeed, some evidence exists in the literature to support the concern that N-terminal tagging can disrupt localization. Using reverse transfection microarrays, Palmer and Freeman (2004) showed that among 16 proteins tested, all those tagged with GFP at the C-terminus were correctly localized in HEK293T cells. In contrast, fewer than half of those tagged at the N-terminus were correctly localized. This result suggests that placing in-frame fusion tags at the N-terminus of a protein might carry more risk than placing tags at the C-terminus. If a C-terminal fusion tag is decided upon, however, then it is imperative to remove the stop codon from the gene of interest and place it after the fusion tag gene.
When you order a tagged ORF clone from GeneCopoeia, our database is linked to data from NCBI and other databases, which includes any known biological data regarding signal peptides. If a protein you are interested in N-terminal tagging is known to carry an N-terminal signal peptide, you will receive a notice that N-terminal tagging of this ORF carries some risk.
Naturally, deciding to place the tag at either the N- or C-terminus based on structural, enzymatic, or localization considerations requires prior knowledge of these characteristics. However, suppose you don’t have a crystal structure, or haven’t yet mapped functional domains of the protein? In that case, we recommend ordering both N- and C-terminally tagged versions of your ORF and testing them independently. GeneCopoeia provides a “secondary clone discount”: When you purchase one version of your ORF clone, you will receive a discount on an alternative version of that same ORF.
Conclusions
GeneCopoeia’s OmicsLink™ Expression-ready ORF clones come not only ready-to-express in your system the same day you get them, but we also verify the DNA sequence. That way, you can rest assured that the protein you are expressing-in either its untagged, native structure, or with a fusion tag-is of the highest quality. Visit our website today to search for your ORF clones of interest, in either untagged configuration or with any of a large selection of fusion tags. Or call us at 1-301-762-0888 to learn more.
References
Bucher, et al. (2002). Differential effects of short affinity tags on the crystallization of Pyrococcus furiosus maltodextrin-binding protein. Acta Crystallographica D58, 392.
Kapust and Waugh (2000). Controlled intracellular processing of fusion proteins by TEV protease. Protein
Expr. Purif. 19, 312.
de Felipe, et al. (2006) E unum pluribus: multiple proteins from a self-processing polyprotein. Trends Biotechnol. 24, 68.
Kozak (2005). A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 33, 6593.
Palmer and Freeman (2004). Investigation Into the use of C- and N-terminal GFP Fusion Proteins for Subcellular Localization Studies Using Reverse Transfection Microarrays. Comparative and Functional Genomics 5, 342.
Ryan, et al. (1991). Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J. Gen. Virol. 72, 2727.
Terpe (2003). Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol and Biotechnol. 60, 523.
HaloTag® is a registered trademark of Promega Corporation. FLAG® is a registered trademark of Sigma-Aldrich Co. LLC.
| Copyright ©2014 GeneCopoeia, Inc. www.genecopoeia.com TNOC1-093014 |