| Ed Davis, Ph.D. |
Recent advances in technologies for genome editing-the use of TALEN or CRISPR to make targeted, permanent changes to genes-have revolutionized molecular genetics. They have also presented users with a choice between these relatively new technologies and that of the more established method of RNA interference (RNAi)-mediated knockdown using short hairpin RNA (shRNA) or short interfering RNA (siRNA). In this Technical Note, we explore the differences between the two methods for ablating gene function, and situations where one technology is more appropriate than the other.
RNAi-mediated gene silencing
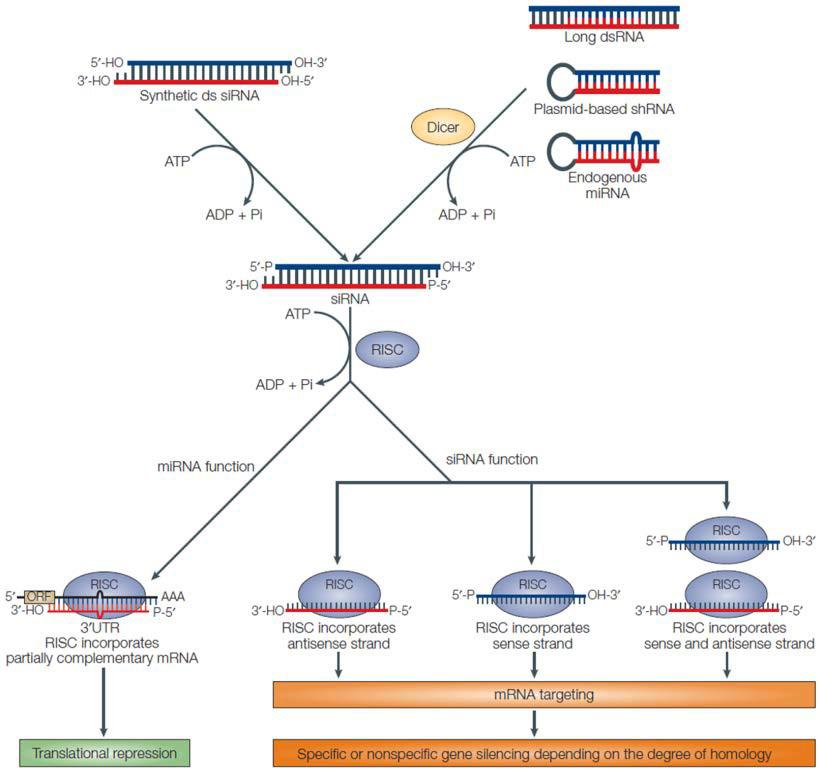
In higher eukaryotes, RNAi-mediated knockdown is the most common strategy for depleting cells of a gene product of interest. However, RNAi usually does not completely shut off the gene. Essentially, short (approximately 20-25 nucleotides) double stranded RNA molecules are either generated from hairpin-forming precursors (shRNAs) or introduced exogenously (siRNAs). After processing by Dicer, a single stranded RNA base-pairs with a target mRNA (Ketting, 2013). Depending on the organism, RNAi- mediated gene silencing is carried out by Argonaute proteins via either mRNA degradation or translation inhibition (Figure 1). The end result is post transcriptional down-regulation of gene expression, without changing the genetic code (Mittal, 2004). Some functional RNA or protein remains and is translated at lower levels. So, the RNAi strategy for reducing gene function is termed a “knockdown”. Gene function is reduced, but not eliminated.
Figure 1. General scheme of RNAi pathways. From Mittal (2004).
Genome editing for gene knockout
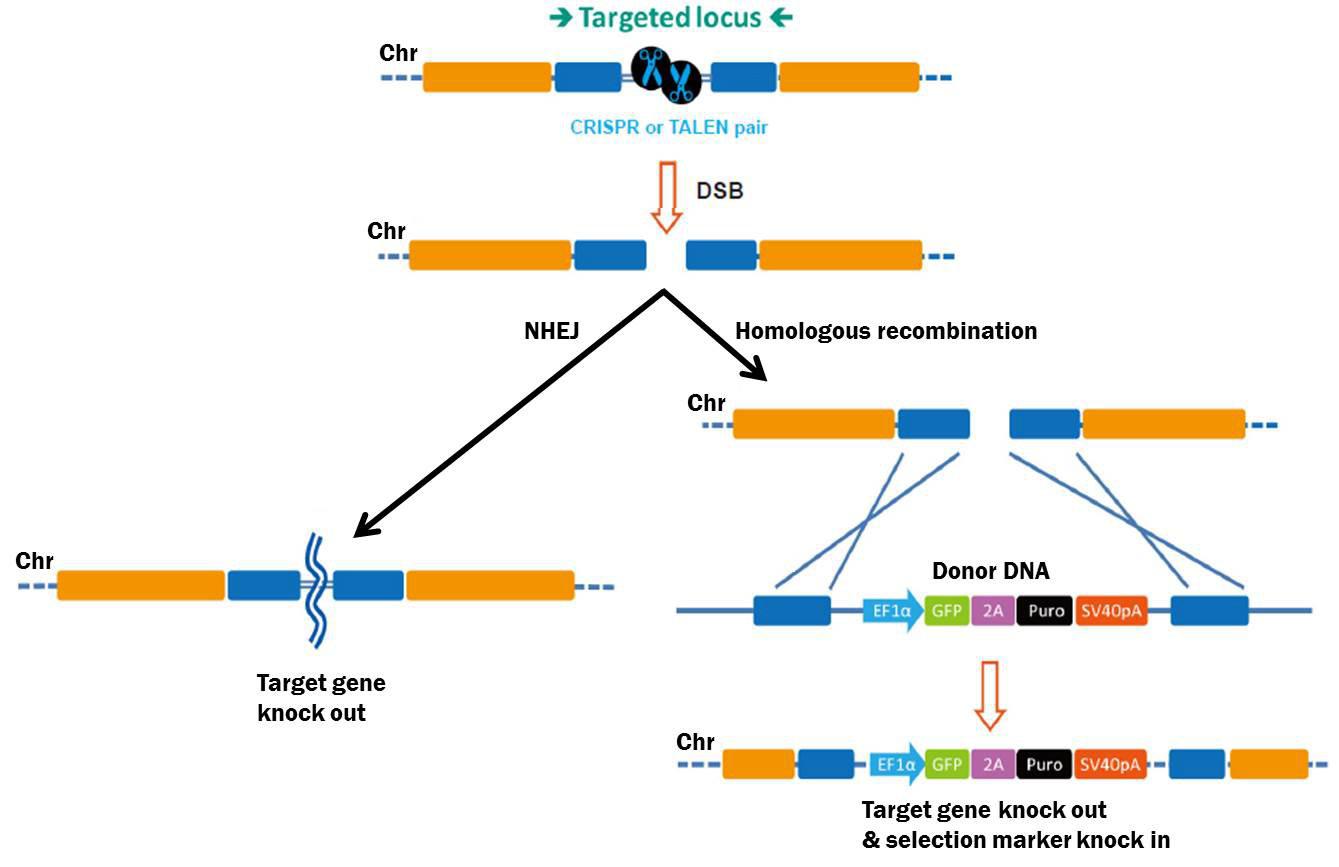
By contrast, genome editing changes the genetic code, typically causing a “knockout”, or complete elimination of gene function. The process begins with creation of a double-strand break (DSB) in the chromosome (Bogdanove & Voytas, 2011). Recently, two tools have been developed for generating DSBs with high efficiency: Transcription Activator-Like Effector Nucleases (TALENs), and Clustered, Regularly Interspaced Palindromic Repeat Associated (CRISPR-Cas) proteins (Bogdanove & Voytas, 2011; Jinek, et al., 2012; Shalem, et al., 2014; Wang, et al., 2014). Both these tools are adapted from bacterial systems that either cause plant pathogenesis (TALEN), or defend the genome from insertional mutagenesis (CRISPR-Cas). TALENs are chimeric proteins consisting of site-specific DNA binding proteins fused to the restriction endonuclease FokI. CRISPR-Cas uses a site-specific, 20 nucleotide single guide RNA (sgRNA) to bring the Cas9 nuclease to its target locus. For both TALEN and CRISPR-Cas, the nuclease cuts both DNA strands of the target. This break must be repaired or the cell will die, so eukaryotic cells respond by two major mechanisms (Figure 2). The first, non-homologous end joining (NHEJ), re-ligates the two free chromosome ends. However, NHEJ is error-prone, often resulting in small insertions or deletions that can disrupt, or knock out, the gene. Alternatively, cells can repair DSBs through homologous recombination (HR), which provides researchers more options for gene knockout. Defined deletions can be introduced, insertional mutations can be created, or single bases can be changed, to name a few.
Figure 2. Pathways for repair of DSBs induced by genome editing tools. Left: Non-homologous end joining. Right: Homologous recombination in the presence of a donor template.
Comparisons between RNAi and genome editing
So, which strategy is better: RNAi-mediated knockdown, or genome editing-mediated knockout? This depends on the experimental goals (Table 1). Indeed, some people confuse the two strategies, referring to genome editing as “knockdown”. This confusion is likely due to the fact that for many years before genome editing became feasible, the most practical strategy for ablating gene function in higher eukaryotes was using RNAi. Thus, researchers became acclimated to the term “knockdown”.
| Method | Knock down |
Knock out |
genetic code |
Change expression level |
Clone isolation required |
| RNAi (shRNA, siRNA) |
√ |
|
|
√ |
|
| Genome editing (TALEN, CRISPR) |
|
√ |
√ |
√ |
√ |
Table 1. Comparison between RNA interference and Genome editing methods for gene ablation.
Basically, RNAi-mediated knockdown is preferable to genome editing when changing the genetic code is undesirable. For example, you might want to reduce gene function temporarily, so you could transiently transfect siRNAs into cells. In a few generations, the siRNAs are lost, restoring normal gene function. Alternatively, you can stably integrate shRNAs into the genome and express them from an inducible promoter. Then you can turn the expression of the gene down and then back up repeatedly, at desired times, and/or in specific tissues. Moreover, shRNA-mediated knockdown does not require the isolation of single clones, unlike genome-editing mediated knockout, so there is less work involved. Finally, completely eliminating gene function might harm the cell, but a partial loss will not.
Alternatively, to make a true genetic null allele, genome editing is preferable (Wang, et al., 2013). Additionally, one might want to introduce a specific point mutation, or correct a pre-existing mutation back to wild type. Finally, you might want to add a fusion tag to your favorite gene and express it from its endogenous locus. These goals absolutely depend on genome editing methods.
GeneCopoeia offers advanced, full-service solutions for both RNA interference and genome editing, from design and construction of shRNAs and TALEN or CRISPR plasmids, all the way up to the generation of stable cell lines and transgenic mice. Please visit our website to learn more: www.genecopoeia.com.
References
Bogdanove & Voytas (2011). TAL Effectors: Customizable proteins for DNA targeting. Science 333, 1843.
Jinek, et al. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816.
Ketting (2011). The many faces of RNAi. Dev. Cell 20, 148.
Mittal (2004). Improving the efficiency of RNA interference in mammals. Nat. Rev. Genet. 5, 355.
Shalem, et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84.
Wang, et al. (2013). TALEN-mediated editing of the mouse Y chromosome. Nature Biotech. 31, 530.
Wang, et al. (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80.
| Copyright ©2014 GeneCopoeia, Inc. Email: inquiry@genecopoeia.com Tel: +1 (866) 360-9531 TNGE1-021814 |