*Promotions are valid in the US & Canada only. For international customers, please contact your
. Promotion ends
. Discounts are not valid on previous purchases and cannot be combined with additional discounts.
Introduction
The All-in-One™ miRNA qRT-PCR reagent kits include the All-in-One™ miRNA qRT-PCR Detection Kits 2.0 and the All-in-One™ miRNA First-Strand cDNA Synthesis Kits 2.0. When used with All-in-One™ Validated miRNA Primers or miProfile™ miRNA qPCR Arrays, the result is high-performance expression-profiling of all miRNAs.
A novel optimized blend of poly A polymerase and reverse transcriptase with an PAP/RT buffer that was specifically developed for the All-in-One Reverse Transcriptase Mix delivers maximum activity of both enzymes for robust first-strand cDNA synthesis from mature miRNAs. The All-in-One™ First-Strand cDNA Synthesis Kit 2.0 is optimized for use with miProfile qPCR Arrays.
The All-in-One™ RT-PCR Detection Kit 2.0 includes both RT and PCR reagents. It combines PCR technology and SYBR® Green to make fast and accurate quantification of mature miRNAs from as little as 10 pg of small RNA or 20 pg of total RNA samples. The kit is designed for use with All-in-One Validated miRNA Primers.
All-in-One™ miRNA assays provides sensitivity and specificity you can afford
- Detect miRNAs from as little as 10 pg of small RNA or 20 pg of total RNA
- Compatible with GeneCopoeia miRNA qPCR arrays or validated miRNA-specific primers
All-in-One™ miRNA assays are fast and easy
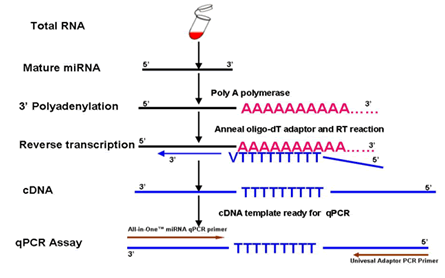
Figure 1. Overview of steps involved in the All-in-One™ miRNA qRT-PCR Detection Kit 2.0. During 3′ polyadenylation, M-MLV RTase and a unique Oligo dT Adaptor primer, reverse transcribes the poly A miRNAs. The kit qRT-PCR Mix, containing SYBR® Green specifically detects the reverse transcribed miRNAs.
Combine these tools with GeneCopoeia miRNA plasmid constructs and assays for comprehensive functional analysis studies.
*miRNA-specific primers sold separately. 
|
|
Trademarks: SYBR® (Molecular Probes)
RT-PCR kits
+ Download miRNA qPCR primer validation report sample (PDF)
FAQs
Frequently Asked Questions: qPCR arrays and reagents
Answer: With these arrays, you don’t need to spend time on primer design and validation of genes and miRNAs. These arrays allow you to perform batch qPCR detection of genes or miRNAs simultaneously in the same 96-well or 384-well plate. This greatly simplifies and speeds up your experiments.
Answer: GeneCopoeia offers custom services for miRNA and gene qPCR arrays that you have designed. You can visit our website and fill out the required information in your order. Please contact us directly if you have any questions.
Custom miRNA qPCR array order form
Custom gene qPCR array order form
Please contact us by email: inquiry@genecopoeia.com or call: 866-360-9531 and 301-762-0888.
Answer: Housekeeping genes are usually used as reference genes, but their expression is not fixed. The expression change of a particular housekeeping gene depends on the experimental conditions. The best method is to choose a series of common housekeeping genes, analyze their expression levels under different conditions, and then select a gene that is stably expressed.
Answer: Your choice of which RT primers to use depends heavily on the experimental design. If you use random primers, reverse transcription will begin at many different sites in the RNA. If you use oligo(dT), reverse transcription will start at the poly-A tail. In quantitative RT-PCR detection of specific genes, we recommend using both random primers and oligo(dT) in one reaction, in order to obtain higher efficiency of reverse transcription. In addition, you can use your own sequence-specific primer to prime reverse transcription in a one-step RT-PCR reaction.
Answer: The DNA polymerase used in these products is a special chemically-modified hot-start enzyme. Incubating for 10 minutes at 95℃ activates the enzyme.
Answer: This kit detects the expression of mature miRNA only.
Answer: The amplification product in miRNA qPCR is about 75bp. The melting temperature of the product is generally between 75℃ and 83℃.
Answer: Most of the GeneCopoeia All-in-One™ miRNA primers are designed to recognize the 3’-end of a template sequence. The matrue miRNA and the pre-miRNA have different sequences at the 3’-end of the base sequence. Therefore, the GeneCopoeia All-in-One™ miRNA primers will not amplify the pre-miRNAs.
Answer: The experimental procedure includes two major steps (Figure 1). In the first step, polyA polymerase is used to add a polyA tail to the 3’ end of the miRNA. Simultaneously, MMLV reverse transcriptase, using a unique oligo(dT) adaptor primer, reverse transcribes the miRNA from the polyA tail. The second step, using the miRNA-specific forward primer and the Universal Adaptor primer as a PCR primer pair, along with a qPCR master mix containing SYBR® Green, specifically detects the reverse transcribed miRNA.
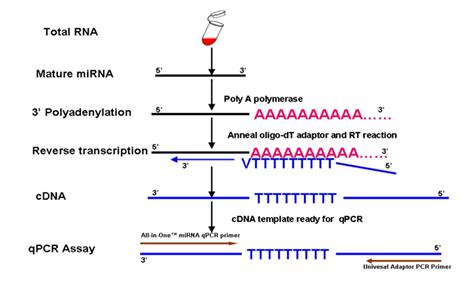 Figure 1. Experimental design used by the GeneCopoeia All-in-One™ miRNA qRT-PCR Detection Kit
Figure 1. Experimental design used by the GeneCopoeia All-in-One™ miRNA qRT-PCR Detection Kit
Answer: A cDNA pool, containing reverse transcribed products from 10 human tissue total RNA samples, is used as the validation template. qPCR was performed using 0.2 uM primer and the GeneCopoeia All-in-One™ qPCR Mix. The GeneCopoeia All-in-One™ qPCR pimers (or the GeneCopoeia All-in-One™ miRNA qPCR primers) are validated to generate a single amplicon of the correct size for the targeted genes and miRNAs and to yield a single dissociation curve peak.
Answer: In combination with the GeneCopoeia All-in-One™ miRNA primers, the GeneCopoeia All-in-One™ miRNA qRT-PCR Detection Kit can detect miRNAs from as little as 10 pg of small RNA or 20 pg of total RNA.
Answer: PolyA polymerase adds poly-A tails to the 3’ end of all miRNAs, which can be reverse transcribed to first strand cDNA in the same tube. The resulting product can be used to detect the expression of any desired miRNAs by qPCR. By contrast, stem-loops are designed to detect just one or a few specific miRNAs, the 3’ end bases of which are paired with their sticky ends. Therefore, to detect such miRNAs, you must select a matching stem-loop. More than one RT reaction may need to be performed at the same time.
Answer: Total RNA has more types of RNA than small RNA, and so provides more templates to amplify in a qRT-PCR detection assay. This can help to improve the amplification sensitivity but could also reduce the amplification specificity.
Answer: The “RT” well contains a spike-in reverse transcription control, which can be used to monitor the efficiency of the reverse transcription reactions. The primer pair pre-loaded in the RT wells specifically amplifies a control cDNA template reverse transcribed from the spike-in exogenous RNA in the sample. The “PCR” well is a positive PCR control, which is used to verify the PCR efficiency by amplifying a pre-loaded DNA template with a specific pre-loaded primer pair. If the RNA sample is of high quality, the cycling program has been correctly run, and the thresholds have been correctly defined, the Ct value of the “RT” well should be 20±3, and the Ct value of the “PCR” well should be 20±2 across all arrays or samples.
Answer: The “GDC” well in each GeneCopoeia ExProfile™ gene qPCR array is a genomic DNA control. This is used to detect the presence of any genomic DNA contamination in each sample during each qPCR reaction. A Ct value for the genomic DNA control of lower than 35 indicates the presence of a detectable amount of genomic DNA contamination. Be sure to remove as much genomic DNA and residual contamination from your RNA samples as possible.



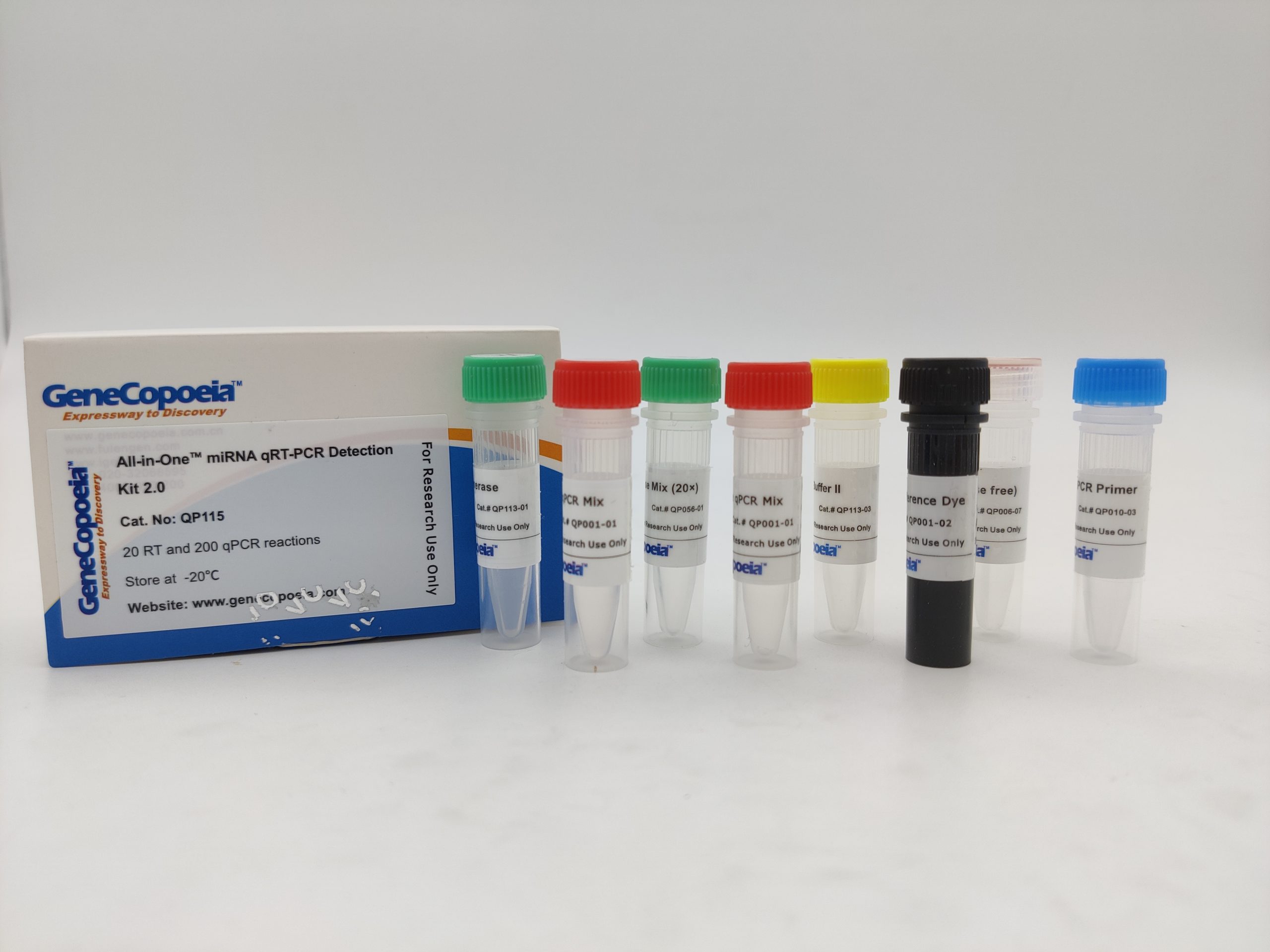


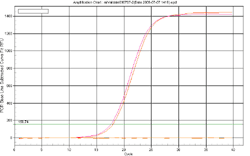
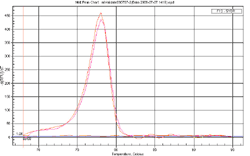
 Figure 1. Experimental design used by the GeneCopoeia All-in-One™ miRNA qRT-PCR Detection Kit
Figure 1. Experimental design used by the GeneCopoeia All-in-One™ miRNA qRT-PCR Detection Kit