Product Information
GeneCopoeia’s IndelCheck™ CRISPR insertion or deletion (indel) detection system provides a powerful means for assisting you in your genome editing applications. The IndelCheck™ system can be used for:
- CRISPR sgRNA functional validation (Figure 1) before undertaking long genome editing projects (3-6 months for genome edited cell lines, or 6-9 months for genome-edited mouse lines), saving you a great deal of time and effort by eliminating CRISPR sgRNAs with poor cleavage efficiency.
- Screening cell clones for knockout (KO) and knock-in (KI) modifications (Figure 2).
Advantages
- Complete system to simplify your CRISPR validation and edited clone screening
- Robust amplification for the target site PCR. No genomic DNA isolation is required
- Easy to use T7 endonuclease I assay with optimized conditions and positive control
Order Information
Order Information
The IndelCheck™ system consists of the following major components:
- Target site PCR kit V2.0. For amplification of targeted genomic regions from cell lysates without genomic DNA isolation. Now in Version 2.0, the Target Site PCR kit is an contains buffer, SuperHeRo™ DNA polymerase, and nucleotides together in one mix for greater convenience.
- T7 endonuclease I assay kit. For detection of CRISPR-introduced indel mutations near target site(s).
- Smart-Join™ Blunt-end PCR Cloning Kit. For cloning of PCR products to be used for sequencing and identification of CRISPR-mediated genomic modifications.
Technology overview
Technology Overview
Application 1: CRISPR sgRNA Functional Validation
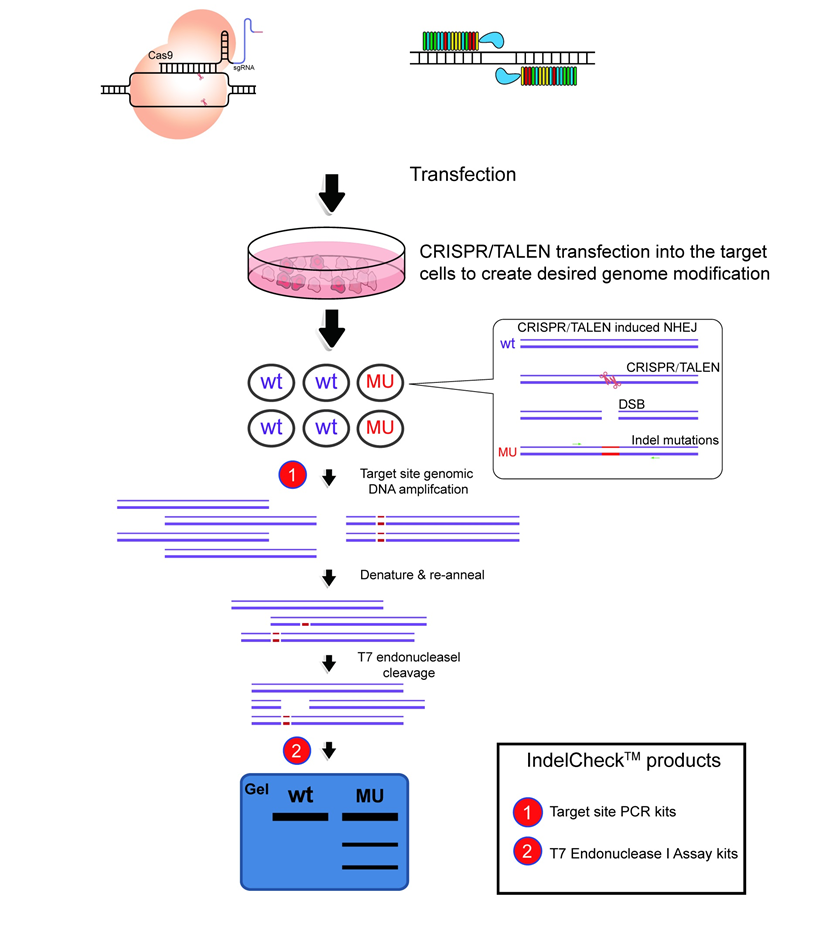
Figure 1. CRISPR functional validation using the IndelCheck™ system. Cells transfected with CRISPR plasmids are harvested in bulk, followed by generation of a PCR product using primers flanking the target site with the Target site PCR kits (1). The PCR product is denatured, followed by re-annealing, leading to a population of double strand fragments, some of which contain mismatches. These mismatches are detected by the T7 endonuclease I Assay kit (2). If CRISPR are active in the cell, then cleavage products will be visible on an agarose gel.
Application 2: Screening for CRISPR-mediated genomic modifications
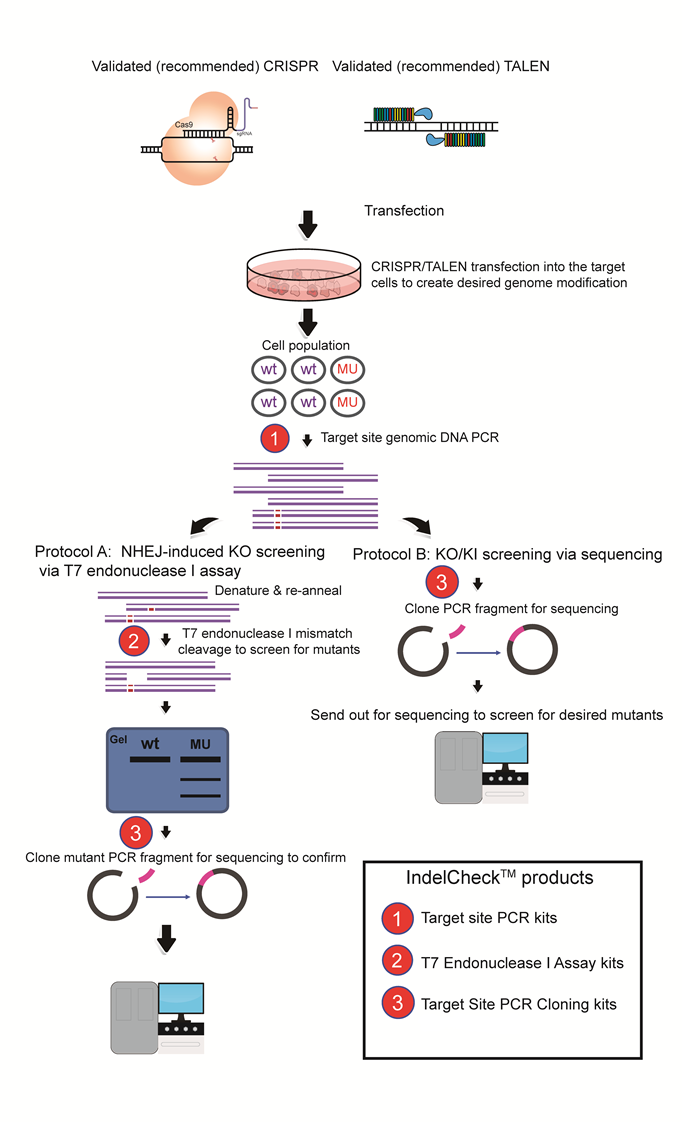
Figure 2. Using the IndelCheck™ system to screen for cell clones carrying desired CRISPR-mediated genomic modifications. Cells transfected with CRISPR plasmids are plated for single clones, followed by genration of a PCR product using primers flanking the target site with the Target site PCR kits (1). In Protocol A (left) for identifying NHEJ-mediated knockouts, PCR products are screened for indels using the T7 Endonuclease I Assay kits (2). Positive PCR products are then cloned into a plasmid vector using the Smart-Join™ Blunt-end PCR Cloning Kit (3) and sequenced to confirm the presence of the mutation(s). In Protocol B, for identifying any knockout or knockin modification, PCR products are cloned directly into a plasmid vector using the Target Site PCR Cloning kit (3) and sequenced to detect the presence of the mutation(s).
Product details
Product Details
Figures showing typical validation results using the combination of IndelCheck™ Target Site PCR kit, T7 Endonuclease I Assay Kit and Target Site PCR Cloning Kit:
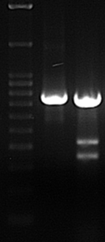
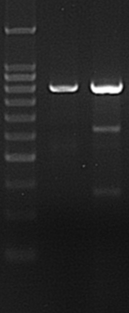
Figure 3. T7 Endonuclease I digestion of genomic amplicons modified by Cas9-sgRNA. Control cells (-) should only have a single band corresponding to uncut amplicon. Amplicons from sample cells (+) transfected with active Cas9-sgRNA yield 3 bands: 1 unmodified + 2 cleavage products of predicted sizes.
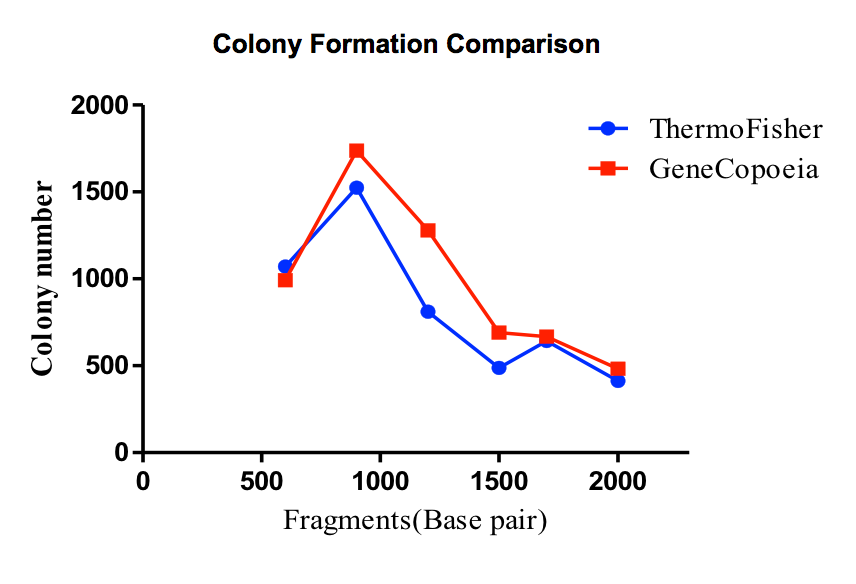
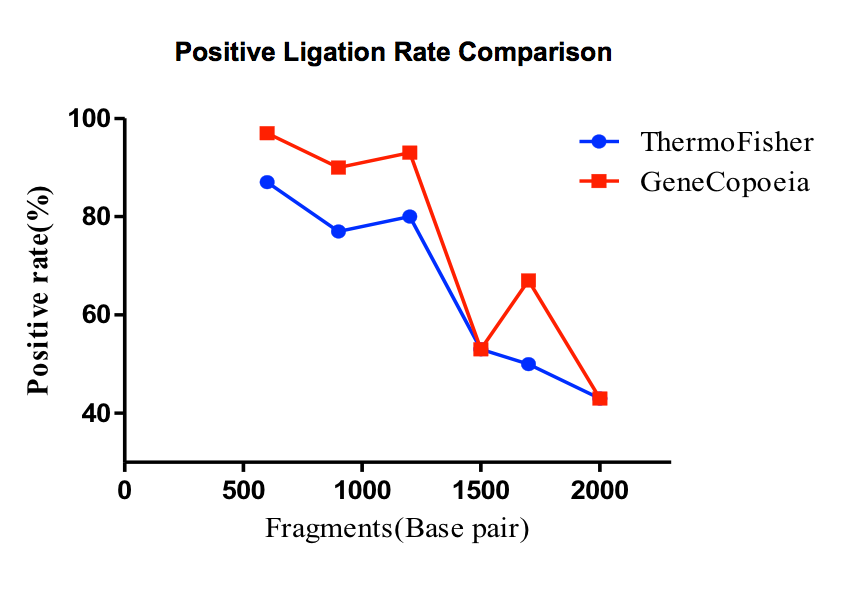
Figure 4. Smart-Join™ Blunt-end PCR Cloning Kit Performance Comparison. A variety of PCR products of different lengths were subjected to ligation reactions and transformed onto agar plates, the total number of colonies and the positive ligation rate were detected. It is shown that the ligation efficiency of Smart-Join™ Blunt-end PCR Cloning Kit (GeneCopoeia, Cat.No IC007) is comparable to that of Zero Blunt™ PCR Cloning Kit (Thermo Fisher, Cat.No K275020).
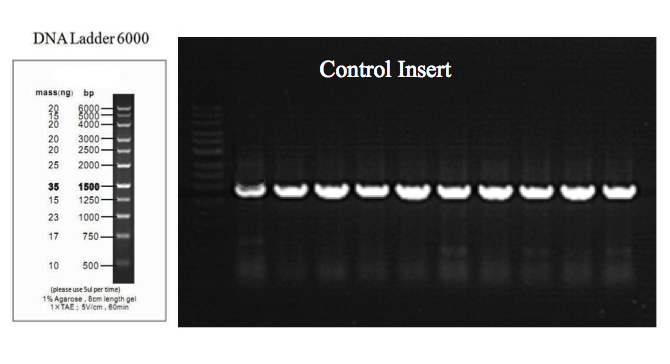
Figure 5. Agarose gel electrophoresis of ligation products from multiple sets of Control Insert using the Smart-Join™ Blunt-end PCR Cloning Kit.
Related products
Related Products
Validation service
GeneCopoeia also offers CRISPR validation services using the IndelCheck™ CRISPR insertion and deletion detection system. We can validate the CRISPR you purchased from us using HEK293 cells (human), NIH3T3 cells (mouse), or your specific cell line(s). Please contact us at inquiry@genecopoeia.com or 301-762-0888.
CRISPR services
GeneHero™ CRISPR products and services