 |
Qihong Xu, Meng Zhang, Xueming Xu, and Ed Davis |
Introduction
Immortalized mammalian cell lines, while providing convenient model systems for biomedical and pharmaceutical research, often carry 3 or more copies of a chromosome or gene (Wistuba, et al., 1998; Burdall, et al., 2003; van Staveren, et al., 2009). For example, the commonly-used human embryonic kidney cell line HEK293 is hypotriploid, with a modal chromosomal number of 64. Further, the ploidy of HEK293 and some other cell lines is not uniform among cells in a population. This presents special challenges for using the clustered, regularly interspaced, short palindromic repeats (CRISPR) system for genome modification in polyploid cell lines in applications that demand complete removal of the endogenous gene product. Thus, the refinement of screening methods to include gene copy number determination would be highly beneficial for genome editing in cultured mammalian cells.
Fluorescence in situ hybridization (FISH) traditionally has been used for chromosome and gene copy number determination in non-genome editing applications, including disease diagnosis and prognosis (Landstrom & Tefferi, 2006; Sarrate, et al., 2010). GeneCopoeia recently released VividFISH™ probes that provide very bright fluorescence in multiple colors (emission wavelengths). In this Application Note, we demonstrate the use of GeneCopoeia’s VividFISH™ probes as an accompanying technique for identifying cells carrying a CRISPR-mediated, penta-allelic knockout of the HDAC6 gene in a cell line carrying 5 copies of the X chromosome. Our results show that including GeneCopoeia’s VividFISH™ probes as part of a CRISPR-mediated genome editing workflow greatly facilitates the post-transfection screening work.
Results
We began a project to completely knock out the histone deacetylase 6 (HDA6) gene (NCBI geneID:10013), located on the X chromosome (Xp11.23), in the human synovial fibroblast cell line MH7A. Mindful of the fact that some cell lines contain more than the normal gene copy number of 2, we were unaware of the number of copies of HDAC6, or of the X chromosome, in MH7A cells. Therefore, we decided to use X- and Y-chromosome-specific VividFISH™ centromere probes to determine the number of copies of the X chromosome in these cells, to help us screen for MH7A-derived stable cell lines containing all copies of the HDAC6 gene knocked out. Using these VivdFISH™ probes on MH7A cells, as well as the diploid male cell line HT1080 as a control, we determined that most of the MH7A cells display 5 fluorescent X-chromosome signals and no Y-chromosome specific signals (Figure 1). In contrast, diploid HT1080 cells display only one X-chromosome signal and one Y-chromosome signal (Figure 1). These results suggest that MH7A cells carry five copies of the X chromosome.
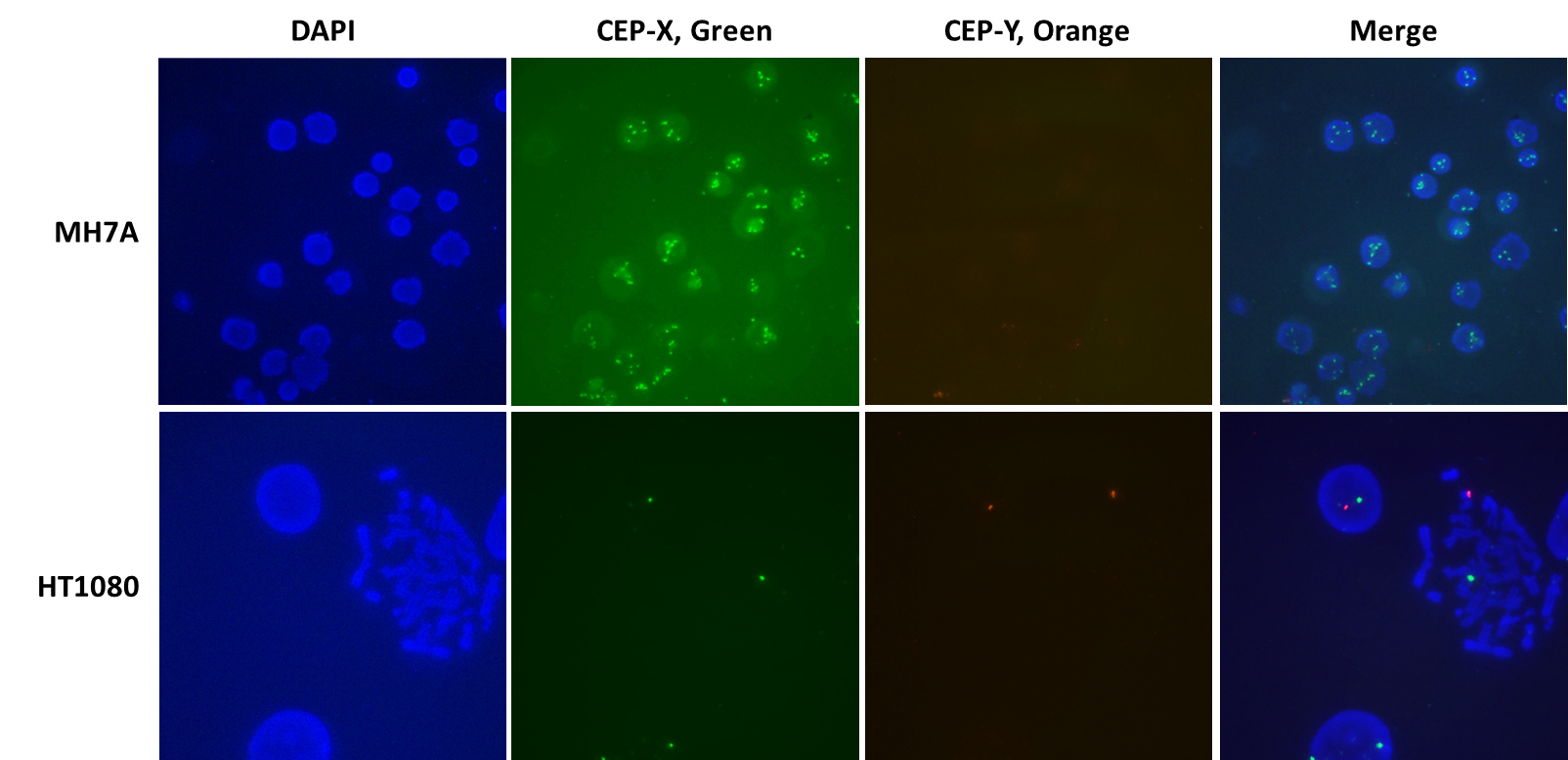
Figure 1. Using VividFISH™ centromere probes to determine the number of copies of the X chromosome in MH7A cells. Unmodified MH7A cells (top) and the diploid human cell line HT1080 (bottom) were fixed on glass microscope slides and hybridized with VividFISH™ probes recognizing the X chromosome (green; GeneCopoeia catalog number FP111), and the Y chromosome (orange; GeneCopoeia catalog number FP117). Only the male cell line HT1080 hybridizes with the Y chromosome probe. Nuclei were stained with DAPI (blue).
For the HDAC6 knockout, we transfected MH7A cells with a plasmid that expresses both the Cas9 nuclease and a single guide RNA (sgRNA) targeting near the initiator ATG of the protein-coding region of HDAC6. Under these conditions, double strand breaks (DSBs) in the chromosome mediated by the Cas9:sgRNA complex are expected to cause insertions or deletions (indels) that usually result in frameshift mutations.
Following transfection, we isolated MH7A-derived stable cell lines grown from single cells. In order to reduce the number of single cell line clones to screen for indel evens, these lines were screened using GeneCopoeia’s T7 Endonuclease I IndelCheck™ assay (data not shown).
The IndelCheck™ T7 Endonuclease I assay kit, based on the mismatch detection assay (Qiu, et al., 2004), generates a PCR product that contains the indel mutation. Therefore, PCR products from the MH7A-derived cell lines carrying indels were subcloned in E.coli, and multiple E.coli subclones from each MH7A-derived line were subjected to DNA sequencing. Analysis of the DNA sequencing data revealed MH7A-derived lines isolated following CRISPR-mediated genome editing that carried from one to five indel mutations. The sequencing results of two lines, each carrying five distinct frameshift-causing deletion alleles, are shown in Figure 2. These results indicate that MH7A cells carry at least 5 copies of the HDAC6 gene, consistent with the VividFISH™ results shown in Figure 1.
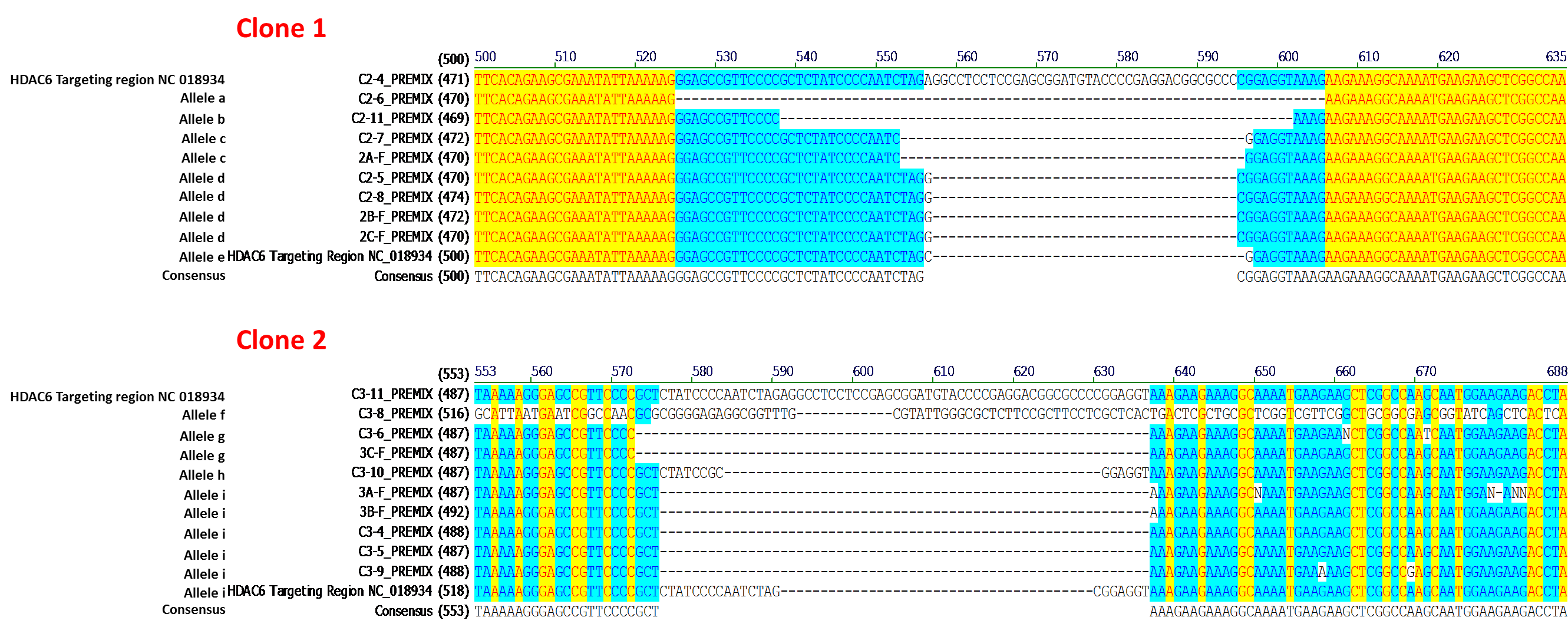
Figure 2. Sequence analysis from two independently-isolated stable cell lines following transfection of MH7A cells with GeneCopoeia sgRNA-expressing plasmids targeting HDAC6 (NCBI geneID:10013). PCR products generated using the IndelCheck™ T7 Endonuclease I kit (GeneCopoeia catalog number IC001) were subcloned in E. coli and subjected to DNA sequencing. Each MH7A-derived cell line carries 5 distinct frameshift alleles resulting from a single transfection.
To determine whether indel-containing MH7A-derived cell lines might be carrying additional copies of HDAC6, either modified or unmodified, we generated additional E.coli subclones from the 3 MH7A-derived lines in which we previously identified 5 distinct indel mutations. In all three cases, we were unable to detect additional modified or unmodified alleles of HDAC6 (data not shown). These results indicated that we were able to use CRISPR to achieve penta-allelic knockout in a cell line that contains five copies of the target gene, and that VividFISH™ probes provided valuable tools for helping us to identify these knockout cell lines.
Discussion
The results presented in this Application Note demonstrate that GeneCopoeia’s VividFISH™ probes are powerful, versatile tools for facilitating CRISPR-mediated genome editing in immortalized cultured mammalian cell lines.
One caveat for using centromeric FISH probes to estimate gene copy number is that gene duplication and translocation might not be detected, resulting in an underestimation of gene copy number. However, we are not suggesting that FISH centromeric probes can be used for absolute gene copy number determination. Rather, we recommend using VividFISH™ results to estimate target chromosome copy number to help estimate the number of cell line clones to isolate for screening for genome editing modifications. DNA sequencing results from edited clones can be used in conjunction with VividFISH™ results to provide a final determination of gene copy number. Therefore, we recommend adding a VividFISH™ chromosome copy number determination step to the beginning of a CRISPR-mediated cultured cell line genome editing workflow.
References
Burdall, et al. (2003). Breast cancer cell lines: friend or foe? Breast cancer research 5, 89.
Landstrom & Tefferi (2006). Fluorescent in situ hybridization in the diagnosis, prognosis, and treatment monitoring of chronic myeloid leukemia. Leukemia & Lymphoma 47, 397.
Qiu, et al. (2004). Mutation detection using Surveyor™ nuclease. Biotechniques 36, 702.
Sarrate, et al. (2010). Role of sperm fluorescent in situ hybridization studies in infertile patients: Indications, study approach, and clinical relevance. Fertility and Sterility 93, 1892.
van Staveren, et al. (2009). Human cancer cell lines: Experimental models for cancer cells in situ? For cancer stem cells? Biochimica et biophysica acta 1795, 92.
Wistuba, et al. (1998). Comparison of features of human breast cancer cell lines and their corresponding tumors. Clinical Cancer Research 4, 2931.
| Copyright ©2016 GeneCopoeia, Inc. www.genecopoeia.com TNGE8-020516 |
