| Ed Davis, Ph.D. |
Introduction
The Clustered, Regularly Interspaced, Short Palindromic Repeats (CRISPR) system is widely used for editing genes and modulating gene expression, thanks to its ease-of-design, efficiency, specificity, and relatively low cost (Wang, et al., 2016). CRISPR-based tools for gene editing are adapted from anti-viral immunity mechanisms present in many bacterial and archaea species. The most commonly-used CRISPR system to date employs the CRISPR-Cas9 nuclease from Streptococcus pyogenes (SpCas9). SpCas9 is a DNA endonuclease that, in the presence of a chimeric single guide RNA (sgRNA), makes a double strand break (DSB) in chromosomal DNA (Jinek, et al. 2012). DSB generation is the necessary first step for most CRISPR -based applications. Alternative variations of SpCas9, such as a nuclease-dead version fused to transcriptional effectors or base editors, Cas9s from other species, or nucleases that cleave RNA, are also utilized for genetic modulation.
The choice of delivery method of Cas9 enzyme and its associated single guide RNA (sgRNA) to mammalian cells or animal models is a critical consideration for scientists when planning gene editing workflows. Most delivery methods work well, but issues such as plasmid integration and off-target effects can have profound influences on experimental results. Delivery of Cas9 protein has several advantages over other delivery methods, including lack of genome integration and lower incidence of off-target modification. In this Technical Note, we discuss the pros and cons of each CRISPR-Cas9 delivery method and introduce GeneCopoeia’s GeneHero™ Cas9 protein for high-efficiency genome editing applications with minimal off-target activity
Cas9/sgRNA delivery methods
The Cas9 nuclease can be introduced into cells or animals as either DNA, RNA, or protein, and each of these formats is amenable for delivery by transfection, electroporation, or microinjection. In addition, viral infection is often used for DNA delivery.
For DNA delivery, Cas9 is typically carried on a plasmid, and is expressed under control of a promoter, similar to the example shown in Figure 1. When Cas9 DNA is co-expressed with sgRNA, gene editing can occur at efficiencies exceeding 70%, as measured by the frequency of insertion/deletion (indel) formation in mammalian cells (Kim, et al., 2014).
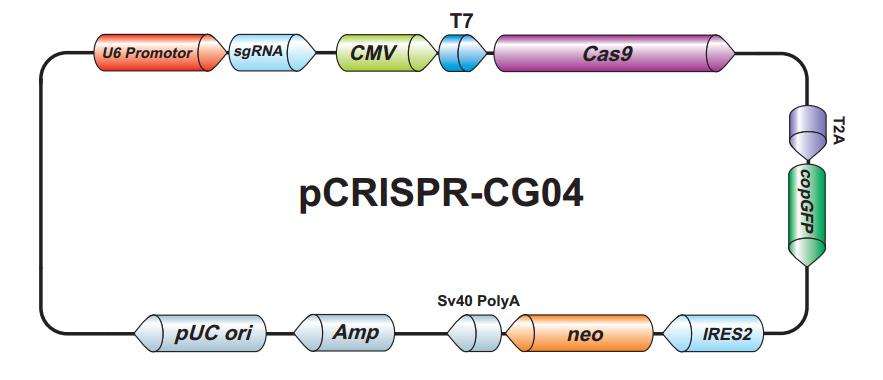
Figure 1. Plasmid expressing Cas9 and sgRNA
Using DNA for Cas9 and sgRNA delivery offers the convenience of having a stable, renewable resource of these reagents, since plasmid DNA can be amplified virtually without limit in bacteria. Indeed, ordering plasmids for DNA delivery of Cas9 and sgRNA is a very popular method of choice for customers at GeneCopoeia. Customers can simply search for their specific gene on our website and choose from multiple vector options. In addition, using lentivirus is a way to deliver DNA to cells that are resistant to transfection, and is an efficient tool for generating stable cell lines expressing Cas9 and sgRNA. GeneCopoeia customers can choose lentiviral plasmids that they can package themselves, or purchase ready-to-use lentiviral particles expressing Cas9 and sgRNA.
Despite the benefits of using DNA for delivery of CRISPR reagents, several papers demonstrate that using Cas9 protein in an RNP complex with sgRNA can provide superior genome editing results.
Jin-Soo Kim’s lab at Seoul National University in South Korea examined both the on-target and off-target editing efficiency of Cas9 protein and compared it with that of plasmid DNA-based delivery (Kim, et al., 2014). Using sgRNA targeting the CCR5 gene produced by in vitro transcription, Kim’s lab electroporated RNP into K562 cells and observed an indel formation frequency of 41%, compared with 50% obtained from DNA. In addition, in hard-to-transfect cells such as BJ fibroblasts and H9 embryonic stem (ES) cells, these investigators noted that on-target editing efficiency for RNP was superior to that of plasmid, with an average indel frequency of 21% caused by Cas9-sgRNA RNPs compared with an average of 10%.for plasmids.
Kim’s lab also observed that ES cells, when transfected with Cas9:sgRNA RNPs, gave rise to about twice as many colonies as those transfected with plasmid DNA, suggesting that RNP transfection is lass toxic to these cells than DNA transfection. However, the reasons for this apparent difference in toxicity are unknown.
In addition to its on-target efficacy, SpCas9 can also cleave at off-target sites, due to its tendency to tolerate non-Watson-Crick base pairing between the sgRNA and the target DNA (Fu, et al., 2013; Kleinstiver, et al., 2016; Slaymaker, et al., 2016; Vakulskas, et al., 2018). Off-target cleavage can lead to unwanted mutations at these locations. Kim’s group compared the off target activities of plasmid- and protein-based Cas9 delivery, and found that, for 3 different sgRNAs, the percentage of indel mutations at four predicted off-target sites was 2-10 fold lower when Cas9 was delivered via RNP than it was for plasmid (Kim, et al., 2014). Because they observed that Cas9 delivered via RNP edits sites much faster and is degraded more rapidly that Cas9 delivered by plasmid, the authors concluded that the lower off-targeting frequency they observed for Cas9 RNPs can be explained by its decreased persistence in cells.
Using Cas9 RNP for gene editing in mammalian cells
To achieve genome editing in mammalian cells using Cas9 protein, it’s essential to provide the sgRNA in RNA format, rather than from a plasmid, because Cas9 protein alone does not contain sufficient charge to cross the plasma membrane of cells (Kim, et al., 2014). Typically, investigators produce sgRNA by either in vitro transcription, using either a plasmid or PCR product expressing sgRNA from the bacteriophage T7 promoter, or by de novo RNA synthesis.
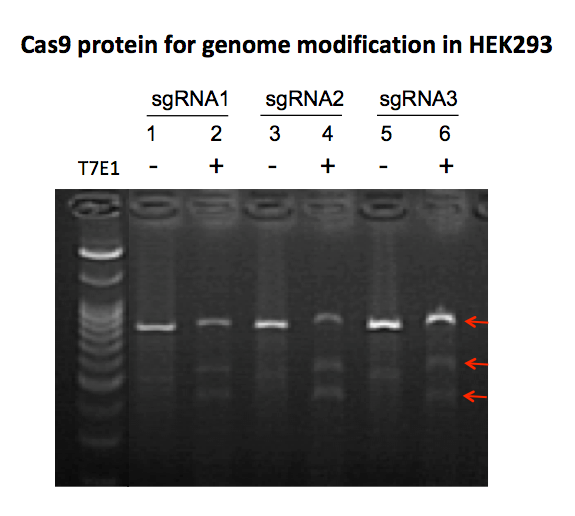
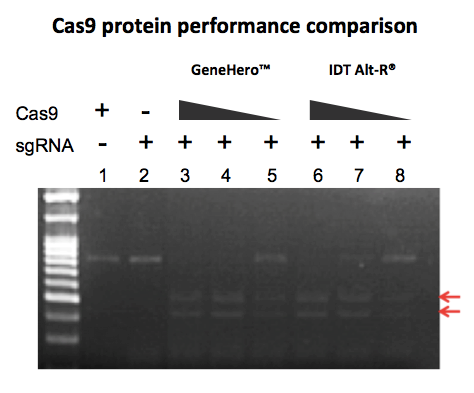
GeneCopoeia’s GeneHero™ Cas9 nuclease is a recombinant protein purified from E. coli. The nuclease carries a nuclear localization signal (NLS), which is required for achieving Cas9-mediated DSBs in mammalian cells. The GeneHero™ Cas9 nuclease is also available either with or without a 6xHis tag. Both versions of the GeneHero™ Cas9 nuclease, when complexed with sgRNA, are capable of efficient gene editing in cell lines, as demonstrated in Figure 2
Additional benefits for using Cas9 protein for gene editing
Aside from its potential for higher on-targeting efficiency and lower off-target modification compared with that of plasmids, using GeneHero™ Cas9 nuclease protein for gene editing in mammalian cells can provide other advantages, such as:
• Lack of genome integration. The GeneHero™ Cas9 nuclease is a protein, so it cannot integrate into the genome. On the other hand, delivery of Cas9 and sgRNA using plasmids or lentivirus often leads to integration. Depending on the application, genomic integration might not be an issue, and in fact might even be desirable. However, there will be other situations in which researchers might want to avoid integration, since integration occurs randomly and can have unwanted consequences on the interpretation of results.
• Faster kinetics. Plasmids encoding Cas9 must be transcribed into mRNA upon entry into the cell. The resulting mRNA is then translated into protein, which must then form an RNP complex with sgRNA. The GeneHero™ Cas9 nuclease-sgRNA RNP complex is made before transfection, so when it enters the cell it is immediately available for genome editing. Therefore, editing can occur much earlier compared with plasmid-based delivery of Cas9 nuclease, which could be useful in some cases.
Using Cas9 RNP for DNA cleavage in vitro
n addition to editing genes in cultured mammalian cells or in live animals, Cas9 protein can also be used for cleaving DNA in vitro, in a fashion similar to that of restriction endonucleases (Anders and Jinek, 2014). This is especially useful for quick validation of synthetic or in vitro transcribed sgRNAs, as seen in Figure 3.
At GeneCopoeia, our Genome Editing scientists have a wealth of expertise with CRISPR applications in mammalian systems. We start at CRISPR design and deliver sequence-verified plasmid DNA or lentiviral particles. We also offer functional validation kits and services for your CRISPR constructs, generate stable cell lines or transgenic mice containing your CRISPR-mediated modification of interest, and provide scientific consulting services to help you plan your projects.
Want to know more about GeneCopoeia’s CRISPR products or to place an order? Visit our website, call 1-866-360-9531, or email inquiry@genecopoeia.com.
References
• Anders and Jinek (2014). In vitro Enzymology of Cas9. Methods in Enzymology 546, 1.
• Fu, et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature Biotechnology 31, 822.
• Jinek, et al. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816.
• Kim, et al. (2014). Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Research 24, 1,012
• Kleinstiver, et al. (2016). High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490.
• Slaymaker, et al. (2016). Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84.
• Vakulskas, et al. (2018). A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nature Medicine 24, 1,216.
• Wang, et al. (2016). CRISPR/Cas9 in Genome Editing and Beyond. Ann. Rev. Biochemistry 85, 227.
• Zuris, et al. (2015). Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nature Biotechnology 33, 73.
| Copyright ©2019 GeneCopoeia, Inc. Email: inquiry@genecopoeia.com Tel: +1 (866) 360-9531 TNGE9-051019 |